The rapid spread of respiratory distress syndrome-2 virus (SARS-CoV-2) worldwide has led to the rapid development of highly sensitive and specific diagnostic tools. These test instruments are largely based on reverse transcriptase-polymerase (RT-PCR) chain reaction to detect total viral ribonucleic acid (RNA). One limitation of the RT-PCR test is that the relationship between advanced RT-PCR and viral infection is not obvious as the disease progresses over time.
Transmission electron micrograph of SARS-CoV-2 virus granules (RA variable B.1.1.7), isolated from patient sample and cultured in cell culture. Image captured at the NIAID Integrated Search Facility (IRF) in Fort Detrick, Maryland. Credit: NIAID
Previous studies have shown that SARS-CoV-2 remains in the upper respiratory tract for approximately 14.5 days after symptoms of disease. However, many studies have also reported that the viral RNA has been present for several weeks, i.e., long ago when most people are infectious. For this reason, the Centers for Disease Control and Prevention (CDC) has developed new guidelines for isolation measures.
According to the previous guidance, based on studies of viral infection in clinical samples, the isolation period for people with moderate to severe disease was ten days from the onset of symptoms. For patients with severe disease or immune conditions, isolation for twenty days was recommended. However, the routine studies suggest that some vaccinated patients may remain infectious for weeks, regardless of the symptoms. This observation has exacerbated the difficulty in formulating strategies for loneliness based on disease symptoms.
The lack of a rapid and high-throughput diagnosis system to separate an infected person from a healthy person makes it difficult to stop the spread of disease. The methods available to diagnose infection are time consuming and impractical. Therefore, scientists are interested in finding molecular markers that may be linked to infection. In this case, they have studied total RNA (positive and negative strands) of SARS-CoV-2 and subgenomic RNA (nucleocapsid (N), spike (S), envelope (E), and membrane (M ), and six protein accessory). The subgenomic RNA can be separated from the total RNA by positioning PCR primers respectively. Evaluation of subgenomic RNA kinase during infection revealed the presence of subgenomic RNA in the neck of SARS-CoV-2 patients up to 5 days after onset of symptoms. Very little information is available on the genotype of subgenomic RNA in hospitalized patients with severe COVID-19 infection and in vaccinated patients.
New paper, published on the medRxiv* preprint server, examining the role of subgenomic RNA transcripts in SARS-CoV-2 disease. Earlier, scientists thought that subgenomic RNA could be an infectious marker to monitor for infection. In the present study, RT-PCR was used to amplify the total and subgenomic nucleoprotein (N) gene and envelope gene (E). Nasopharyngeal samples were collected from 190 patients hospitalized in Michigan between March 13, 2020, and June 10, 2020.
Researchers observed a strong correlation between total RNA and subgenomic RNA. They found that the time points from post-onset symptom are similar, and the reduction in infection rate is the same for both total RNA and subgenomic RNA. Prediction of subgenomic copy numbers from whole copy numbers reveals that subgenomic RNA identification does not add additional information about infection in addition to that obtained from a complete copy number alone. Thus, scientists conclude that there would be no additional benefit in the evaluation of subgenomic RNA.
In this research, scientists found that subgenomic negative E RT-PCR binds positively to time when patients are not infectious. Thus, subgenomic E is not a valid indicator for predicting virus infection. This study also reveals that the median number of days from onset of symptoms to subgenomic negative N RT-PCR is 25 days. Thus, the use of subgenomic N as an indication for infection would significantly extend isolation and not predict infection. This difference appears to be due to the higher sensitivity levels of subgenomic N compared to subgenomic E..
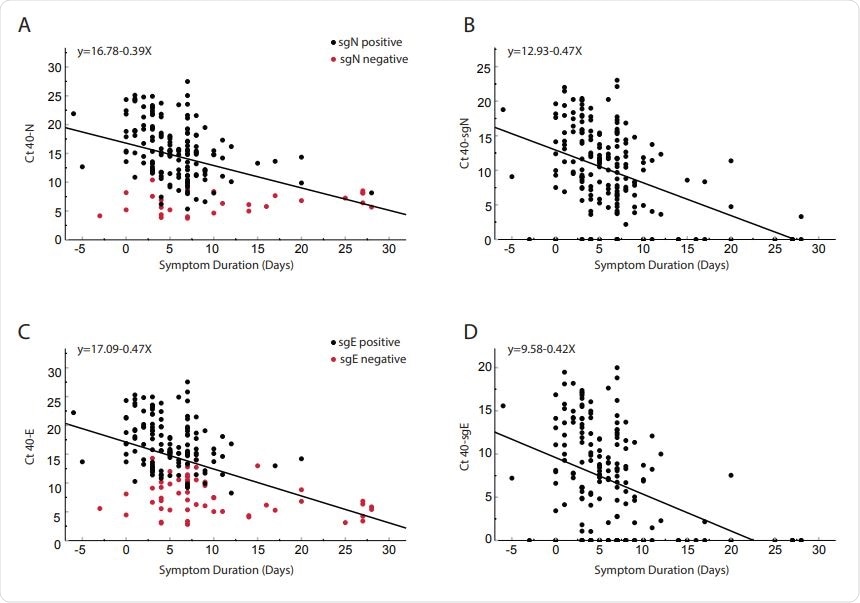
Comparison of day-to-day cycle threshold since symptom onset for clinical samples obtained from 185 patients. Total N (Panel A), subgenomic N (Panel B), total E (Panel C) and subgenomic E (Panel D). Red dots in panels A and C represent subgenomic negative samples and black dots represent subgenomic positive samples. Of the 185 patients, 56 were negative for sgE and 28 negative for sgN (Shown on y-axis). Pearson correlation coefficients: N = 00.404 p <0.0001; SgN = -0.466, p <0.0001; E = -0.456 p <0.0001; sgE = -0.427 <0.0001. Serial reciprocal line equations are indicated in each panel.
Previous research has shown that subgenomic RNA is a suitable marker for active disease because it decays faster than total RNA without reproduction. This result did not align with the findings obtained in the current study. This difference is not due to different contamination of subgenomic transcripts and complete transcripts but due to different detection of transcripts.
The research concludes that infection detection by subgenomic RNA evaluation is not recommended. Similarly, for non-immune patients, the copy number threshold of total RNA provides the same information obtained from the copy number of total RNA. One limitation of the current study is that the scientists have not separated the virus from hospital samples and linked it to a complete or subgenomic copy number. Only a correlation of transcripts to symptom length was used as the conclusion for infection.
* Important message
medRxiv publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical practice / behavior, or be treated as information established.
Magazine Reference:
- Total Viral RNA Load and Subgenomic SARS-CoV-2 in hospitalized patients, Derek E. Dimcheff, Andrew L. Valesano, Kalee E. Rumfelt, William J. Fitzsimmons, Christopher Blair, Carmen Mirabelli, Joshua G. Petrie, Emily T .Martin, Chandan Bhambhani, Muneesh Tewari, Adam S. Lauring medRxiv 2021.02.25.21252493; doi: https://doi.org/10.1101/2021.02.25.21252493, https://www.medrxiv.org/content/10.1101/2021.02.25.21252493v1