Researchers report a method of removing sound from high-resolution transmission electron microscopy images using probabilistic genetic models.
An electronic microscope (EM) is a method used to see viruses and their infectious processes, allowing us to see how it binds to cells and spreads through the body. An electronic transmission microscope (TEM) can produce very high resolution images at the nanometer scale, but these low light images generally have enough noise. Images of viruses often appear flat or 2-dimensional.
Several noise reduction algorithms have helped to improve image quality, allowing better interpretation of the data. For true respiratory coronavirus 2 (SARS-CoV-2) syndrome, EM images were also used to reconstruct 3-dimensional (3D) images of the virus.
This usually involves the use of a large number of TEM images, for example, to generate 3D structures of proteins. A computed tomography study used cryoelectron to record many TEM images of the virus because the sample is inclined on an axis, and the obtained images are integrated by software. If only acoustic images are available, standard approaches such as feed-forward neural networks cannot be used as they require a large number of pairs of clean and noisy images.
Denoising one TEM image
In a new study published on the bioRxiv * preprint server, researchers from Carl von Ossietzky Oldenburg University report a method to obtain 3D images of the virus using a single TEM image.
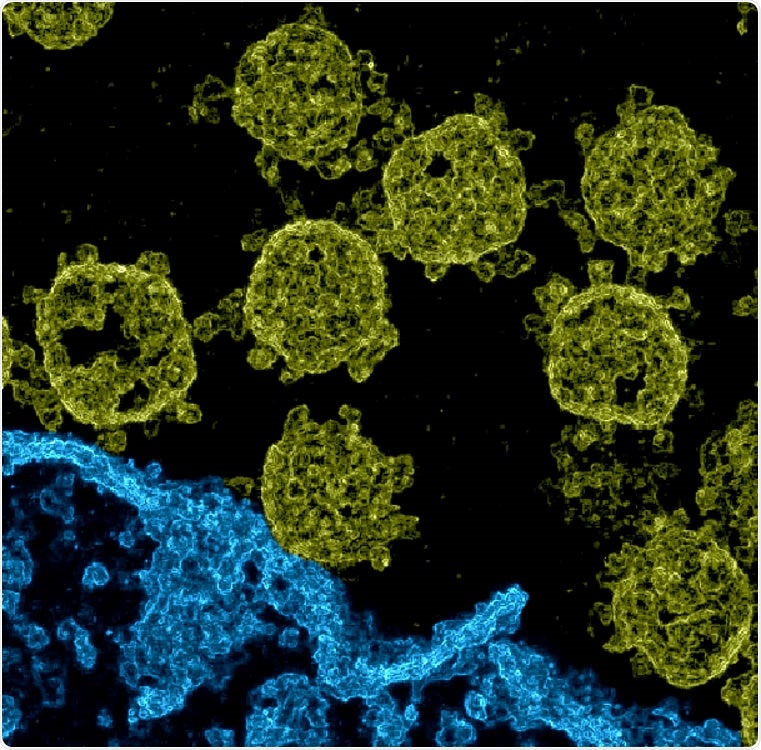
Imaging of SARS-CoV-2 viruses in Vero cell cultures. We used data provided by Laue et al. (2021) used a diffusion electron microscope of ultrathin plastic particles. Based on the data, we estimated pixel differences during the application of probabilistic Machine Learning algorithms for denoising (Sheikh et al., 2014; Monk et al., 2018; Drefs et al., 2020). . The resulting image gives these spatial differences (see also Supplementary Figure 1, which shows a close comparison between a segment of the acoustic EM image and the corresponding approximate pixel variables). The image presented here was obtained after a different elevation and coloring: structures we named by hand belonging to a cell were colored in blue (the lower right-hand place), the excess color in yellow.
They first used probabilistic genetic models to study data representations of the TEM images. These techniques are unstructured and usually require less data to learn the relevant representations. If there is only one image available for learning, image processing is complex. However, sexual image statistics often contain high-quality, usable information.
The researchers used a freely available TEM image of SARS-CoV-2 in Vero cell cultures and used acoustic data in the form of extracts extracted from the image. After cutting the image to a smaller size, they halved the pixel density by removing every second pixel to reduce computing demand. Their approach was to generate the denoised image by estimating the average of each pixel.
They also processed the same image using other probabilistic models. Using the Gamma-Poisson combination model changes the spatial appearance and to a lesser extent the appearance of the normally converted image. The models that provide clear test rotation for image processing are important, but non-local image processing methods were less suitable.
Visual inspection of the denoised image confirmed that the quality was similar to those obtained using modern algorithms and effective sound suppression in high-resolution EM images.
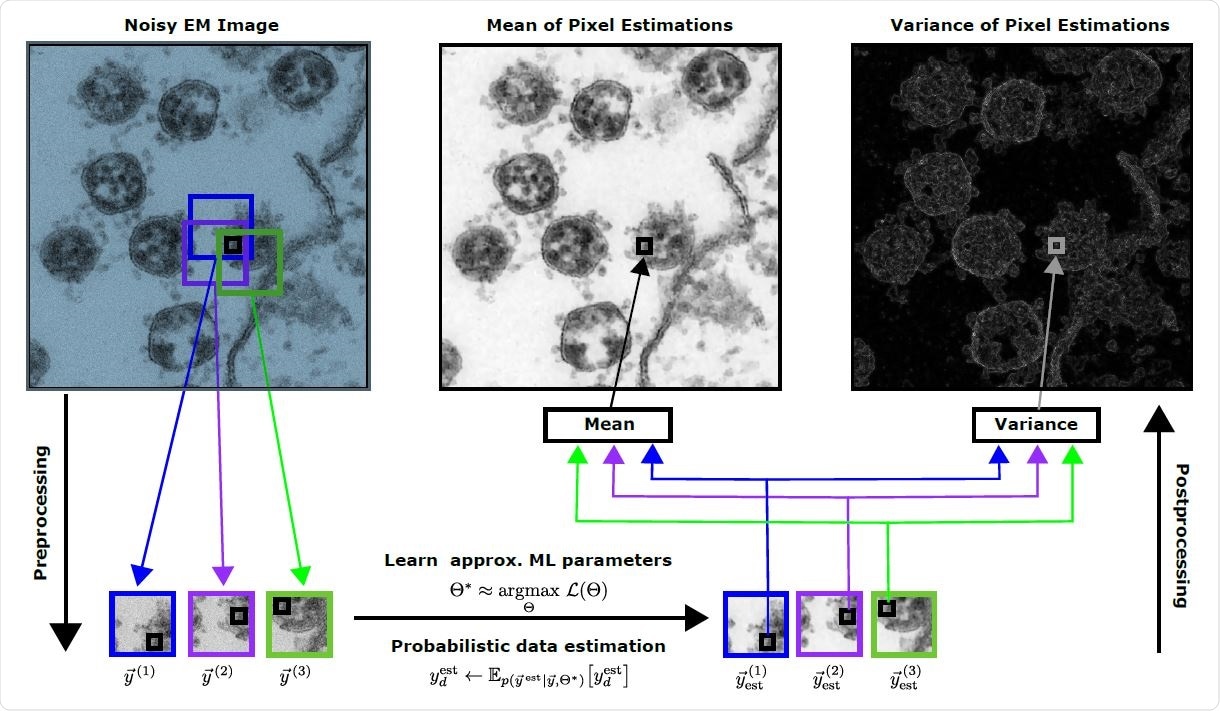
Development of `Zero-Shot ‘on Electronic Microscope Imaging of SARS-CoV-2 viruses using probabilistic Machine Learning algorithms for rejection. The insert (top left) is a version and scale of the Dataset 07 SARS-CoV-2 077.tif file from Laue et al. (2020, 2021). Using patches cut out of the acoustic input, we learn a probabilistic representation of the EM image using a linear spike-and-slab scatter coding model. We can then apply the learned representation to reconstruct each EM image patch to probabil. Finally, we generate reconstructed EM images (middle top and right) with computational methods and variations of the pixel estimates from different folders. The EM image development method does not require training data (clean) and can be sent directly to a single audio image
Advantages and disadvantages
The team also analyzed the reconstruction statistics. The pixel variability estimates can also be used to provide a spatial view of the lesion area. Although reconstruction of previous tomographic images may provide high-quality images, they require many TEM images of a single virus, which is difficult to obtain. This method cannot reproduce 3D images of a complete disease scene.
Each method used for 3D production can incorporate its own art materials. For example, artifacts may be present in the new way because there is less representation of rare structures. Furthermore, because the reconstruction is based on a single image, a real surface is not considered.
An advantage of the new method is that a surface does not interfere with other structures, but a disadvantage is that internal viral structures can interfere with surface structures. Because different methods have different advantages, they can be combined in hybrid systems.
Different approaches are usually used for different data. For example, the previously reported protein reconstitution method combines TEM images of several protein molecules of different viruses, which the new method cannot. The new method cannot combine multiple TEM images of the same virus to generate a 3D image. However, the other methods cannot retrieve a spatial view from a single acoustic image.
Although spatial imaging of virus infections has been reported before using scanning electron microscopy images, they have low resolution and do not show the details of the virus. The images obtained with the new method for the SARS-CoV-2 contain a high level of information, which may lead to a better understanding of the disease.
The original color and image can be downloaded for download at this museum.
* Important message
bioRxiv publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical practice / behavior, or be treated as information established.