A recent study by U.S. researchers shows how the 501Y.V2 variant of acute respiratory coronavirus 2 (SARS-CoV-2) syndrome, characterized by a number of mutations, is able to escape from neutralization by resistance to SARS-CoV- first wave. 2 antibodies and potential relapses of COVID-19. The paper is currently available on the bioRxiv * preprint server.

As many variants of SARS-CoV-2 emerge and then eradicate first-wave viruses, it is crucial not only to evaluate their transmission. and depression in the cause of coronavirus infection (COVID-19), but also the desire to escape antibody neutralization.
Of particular interest are mutations that harbor mutations that may affect the interaction of the viral spike binding domain (S RBD) with the viral receptor on host cells, an enzyme 2 that converted angiotensin (ACE2), which provides an entry point for the coronavirus.
Changes with a more related relationship for ACE2 are likely to spread more. In addition, transmission is associated with mortality, as an inevitable increase in disease rates caused by the novel variables leads to higher disease and death toll.
However, these horrific effects of faster and more widespread diseases can be exacerbated by the loss of effectiveness of current antibody-based therapies and vaccines and a decrease in immune function in individuals previously infected with the virus. ‘first wave’.
To develop our understanding of the risks of individual or integrated mutations in these ‘second wave’ variables, a research group from the California company ImmunityBio conducted a computer analysis of the interaction of the S RBD with human ACE2 .
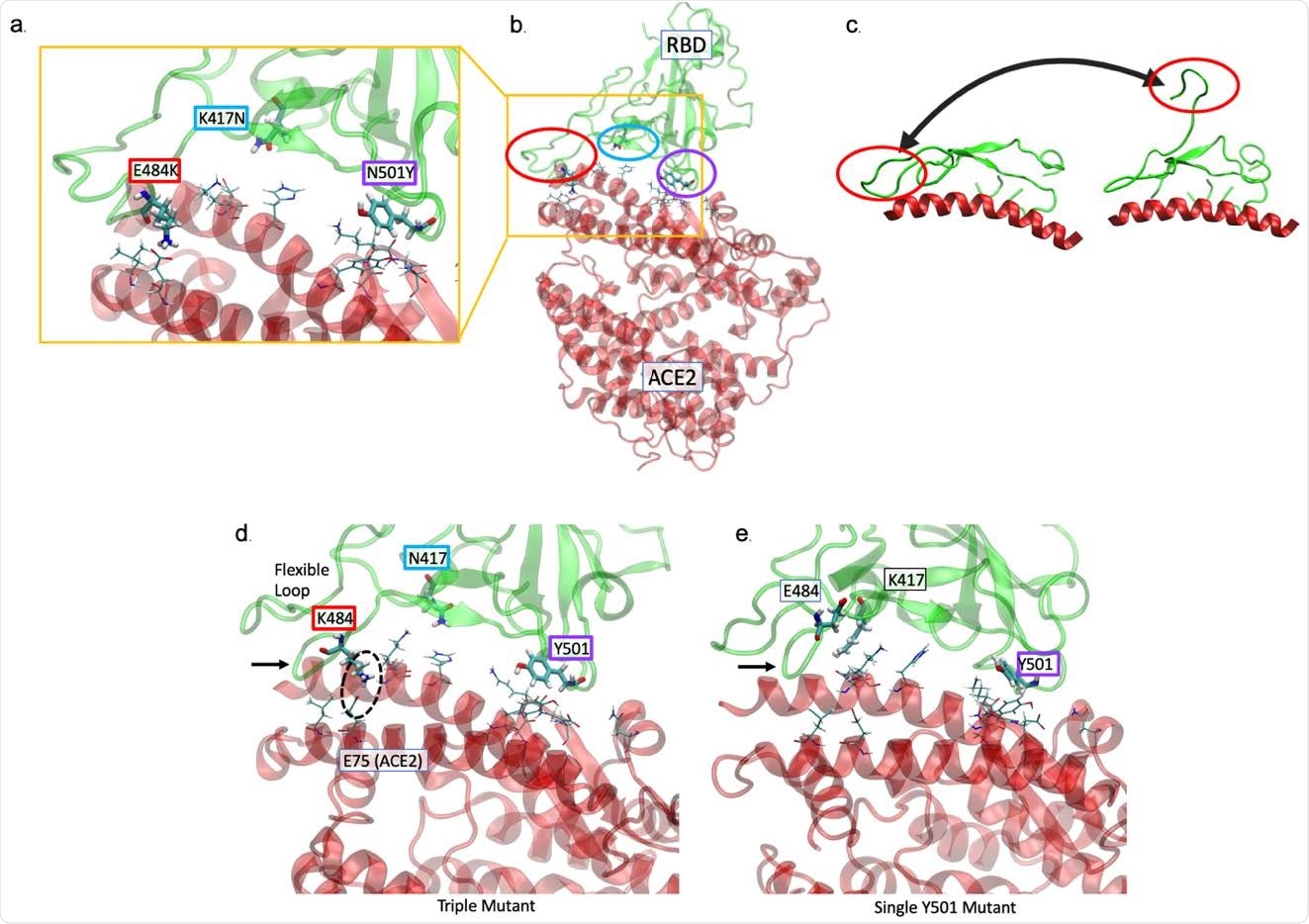
The substitution of K484 in the South African novel novel increases the affinity of the spike-binding domain (S RBD) for ACE2. (a, b) The position of the substitution E484K (red), K417N (cyan), and N501K (purple) is at the interface of the interface 501Y.V2 S RBD – hACE2. the residue of hACE2 closest to the residue of RBD is presented as thin sticks. The E484K mutation is located in a highly flexible loop area of the interface, K417N in an area with a lower contact probability, and N501K at a second point of high affinity connection. (c) The range of motion available with the curve containing residue 484 is shown by PCA of first-wave sequence MD simulation11,13. (d) MD simulations performed in the presence of the 3 surrogates show that the curve area is tightly connected (black arrow) by hACE2. A key pair of communication ions is circulated. (e) In contrast to K484, when E484 (‘wildtype’) is present with only the Y501 difference, the loop is less connected (arrow).
In silico simulation techniques
In this study, the researchers have used millisecond-scale MD simulations to study mutations (E484K, K417N, and N501Y) at the S RBD-ACE2 interface in the 501Y.V2 difference in the fast-spreading South Africa – and their impact on RBD binding affinity and glycoprotein spike conformation.
The wild-type ACE2 / RBD wild-type was extracted from a cryo-electron microscopy structure. In addition, ten copies of each RBD mutant were reduced, balanced and simulated, and the reduction was processed in two stages.
Finally, key component analysis (PCA) was followed using the full set of simulations of the triple mutant systems, E484K and N501Y. Simulation structures were installed in the eigenvectors for each mutation system.
The great escape from nesting
The study revealed a greater affinity of K484 S RBD for ACE2 compared to E484, as well as a greater probability of modified concordance compared to the original structure. This may represent mechanisms by which the new 501Y.V2 viral variant was able to replace the original SARS-CoV-2 rays.
In particular, both E484K and N501Y mutations were shown to have a greater affinity of S RBD for the human ACE2 receptor, while E484K was able to alter the charge on the flexible loop region of RBD, and as a result of the that favorable novel connections were created.
The previously mentioned improved relationship is to blame for the faster spread of this variability due to increased transmission, which is a key reason why it is important to monitor these mutations and work in a timely manner. .
In addition, the entry of conformal mutations is dependent on escape from the 501Y.V2 variant (different from the UK variant B.1.1.7 with the presence of E484K mutation) from neutralization with anti-SARS-CoV-2 antibodies existing and re-infect COVID-19 individuals.
Effects for further vaccine design
“We believe that the MD simulation method used here also represents a tool used in the arsenal against chronic pandemic, as it gives us an insight into the probability mutations alone or in combination that may have side effects that reduce the effectiveness of existing treatments or vaccines “, say the authors of this study.
“We suggest that vaccines whose efficacy relies heavily on humoral responses to the S-antigen alone are partially limited by the emergence of novel strains and rely on frequent remodeling,” they conclude. adds.
On the other hand, a vaccine that triggers a much less active T-cell response is subject to changes as a result of mutations and, therefore, provides a better and more effective means of protection against this disease.
Finally, the excellent vaccine would contain a second preserved antigen (such as the nucleocapside protein SARS-CoV-2), which may elicit a humoral and mediated immune defense response – even when it comes to the against a rapidly changing virus.
* Important message
bioRxiv publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical / behavioral practice, or be treated as information established.