-1.jpg)
The clinical phenotype of coronavirus disease 2019 (COVID-19), caused by the acute acute coronavirus 2 syndrome (SARS-CoV-2), is remarkable for the wide range of concerns among individual patients. Some of these differences are known to be affected by genetic changes.
To investigate these differences, researchers in a new study used human pluripotent stem cells (iPSCs) from different genetic individuals. These cells are used to model genetic disease because they contain the donor’s genetic information.
The study, published as an introduction to the bioRxiv * server, using a panel of iPSCs from over 500 people. The researchers preferred anonymous iPSCs to reduce the time required to differentiate, especially since infection is not always reliable.
Establishment of SARS-CoV-2 infection in iPSCs
Human iPSCs are not infected with SARS-CoV-2 because they express the viral receptor receptor, the enzyme 2 that converted angiotensin (ACE2) at low levels. These cells have high levels of CD147 coronavirus receptors and TMPRSS2 serine protease.
To make them susceptible to SARS-CoV-2 infection, the iPSCs were engineered with the relevant genes, inserted by adenovirus (Ad) vectors, with almost 100% efficacy. These genes encoded human ACE2 and TMPRSS2 at hypertensive levels, leading to severe infection with the virus. Excessive TMPRSS2 transfer alone was not appropriate to achieve this effect.
Two days after infection of the ACE2-positive iPSCs, cell fusion was observed, with cell death after four days, in many cases. Under an electron transduction microscope (TEM), the infectious cells showed character changes, including zippered endoplasmic retopulum (ER), two-membrane spherules (DMS), and viral fragments near the cell membrane.
Viral fragments have been observed within the endoplasmic reticulum-Golgi intermediate region (ERGIC), along with two-membrane vesicles (DMVs.The latter are at the heart of viral RNA synthesis and are absent from unprotected cells.
Therefore, TEM is a useful tool to study the life cycle of this virus.
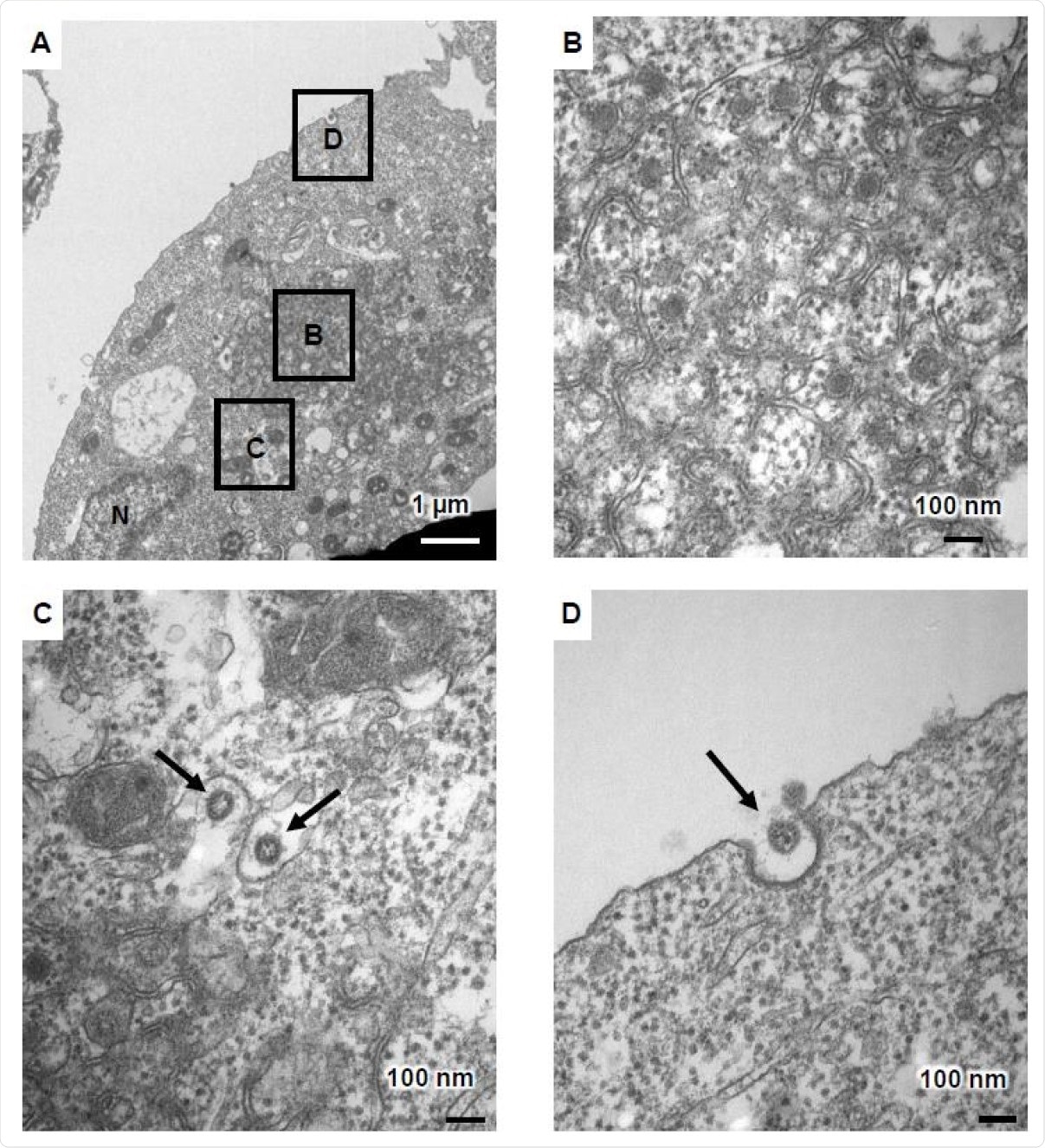
TEM images of infectious ACE2-iPS cells (A) TEM images of infectious ACE2-iPS cells. A zippered endoplasmic reticulum, a double-membrane spherule (DMS) (B), and viral fragments were observed in the ERGIC (black arrows) (C) and near the cell membrane (black arrows) (D).
An involuntary state follows an infection
Gene and protein expression was assessed on day 3 after infection in infectious and non-infectious cells. This showed high levels of viral genome and ACE2 in infectious cells, but not of unknown cell markers or tissue immune markers. The viral protein nucleocapsid (N) was strongly expressed in the infectious cells on day 2 after infection.
RNA-seq analysis of these cells showed that, in approximately 7% of all genes, there was a quadruple or greater change in the level of expression, and none of these encoding traits showed signs of disrespect or texture protection.
Endoderm signals, except the CER1 gene and for CD147, NRP1, and TMPRSS2 genes, called SARS-CoV-2-associated markers, remained unchanged. Mesoderm and ectoderm symptoms were also unchanged.
These findings suggest that human iPS cells maintain an unknown state even when SARS-CoV-2 reproduces in large numbers. ”
Candidate drug assessment
Compared to Vero cells, iPSCs have been shown to show stronger drug effects on SARS-CoV-2 infection, with the exception of interferon-beta. Of the eight drugs tested, remdesivir showed the greatest antiviral activity, and ivermectin showed high cytotoxicity.
Chloroquine and Favipiravir failed to suppress viral reproduction. The RNA-dependent RNA Polymerase (RdRp) inhibitors (Remdesivir and EIDD-2801) and TMPRSS2 inhibitors (Camostat and Nafamostat) were shown to have antiviral activity in these cells, revealing the value in drug evaluation
Individual differences
The researchers then sent iPSCs from eight different donors to study infection differences due to individual factors only. The virus reproduced with different efficacy among the eight donor cell lines, despite the comparative expression of ACE2 in all of them.
The virus showed increased reproducibility in male progenitor cells than females, reflecting the potential for reproducibility differences in reproductive vulnerability in these cells. Both the androgen receptor and the target gene TMPRSS2, the latter of which are expressed at slightly higher levels in male iPSCs, may be related to this difference.
What is the impact?
The viral life cycle can be monitored in these human iPSCs if the ACE2 receptor is overexpressed. The study showed the operation of sex-specific differences in the efficacy of infection and viral reproduction. Drug-blocking effects were also seen in this cell model.
These findings suggest that, using our iPS cell panel, it will be possible to study the effects of race and blood type as well as sex on SARS-CoV-2 infection. ”
In addition, it is possible to identify genetic mutations that occur at high frequencies in cells that maintain infectivity by using iPSCs from a panel with recorded genomic information.
Although the efficacy of infection is different among different donors, the fact that none of these donors lacked COVID-19 limits the conclusion that they were susceptible to the disease. The study is being advanced using iPSCs from patients with mild and severe COVID-19, allowing results in infectious cells to be compared with the patient’s clinical presentation.
Although mutations in some immune genes may be related to the depth of symptoms, further research is needed to understand how these genes work during COVID-19. This can be done conveniently in iPSCs, which allows the easy integration of single nucleotide mutations.
It should be noted that the role of ACE2 overexpression has not been elucidated in the individual differences in SARS-CoV-2 infection within iPSCs from different donors. If so, another system using ACE2-expressing somal cells must be used to first assess the sensory and mimetic effects of ACE2 and other related genes.
Causes of individual, as well as genetic, differences are not addressed by this system, including those due to advancing age and associated reductions in CD8 T cell cytotoxicity. Studies of blood samples could help establish the cause of these differences in viral vulnerability and reproducibility. The ACE2-positive iPSCs remain a useful platform to study and determine the cause of individual changes, identify high-risk groups and evaluate new drugs.
* Important message
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical / behavioral practice, or treated as fixed information.