Extensive testing for the acute respiratory infection coronavirus syndrome 2 (SARS-CoV-2) has been a major mitigating step in the control of chronic pandemic. The disease causes the 2019 coronavirus infection (COVID-19), which to date has claimed more than 2.7 million lives worldwide. The need for effective therapeutic solutions is good, even with the ongoing vaccination programs against SARS-CoV-2 in many parts of the world. One plasma-based therapeutic approach is the use of neutral antibodies from the blood of convalescent patients.
To address the need for a high-quality neutralization assay against SARS-CoV-2, researchers from the National Institute of Infectious and Infectious Diseases (NIAID) modified and optimized a previously established fluorescence-reducing neutralization assay (FRNA) to evaluate qualitatively quantify large numbers of infectious cells through the use of a high-content imaging system.
In this study, the team described a semi-high throughput, highly quantitative neutralization assay for SARS-CoV-2 built around Operetta’s high-content imaging system. The authors noted that similar equipment (which may not be available in many laboratory settings) could be just as effective if properly tested.
They reported that, by February 2021, the SARS-CoV-2 FRNA had been used to screen more than 5,000 samples, including acute and convalescent plasma or serum samples and therapeutic medications, for the neutral titers SARS-CoV-2.
While variations in live virus assessments will always be due to the nature of the biological system, using a rigorous statistical approach to inform data acceptance can have adverse effects. which may have poor infectious efficacy, piping errors, marginal effects, and inconsistent staining. . ”
COVID-19 is a heterogeneous disease, often characterized by severe respiratory disease, cardiovascular disease, and neurological disease. Rapid conversion of antibody class from immunoglobulin M (IgM) to IgG and IgA occurs at the time of true infection. The researchers note that the antibody isotype is important in controlling the disease, as is the target viral protein.
The researchers maintained a link between disease depth and stronger and longer antibody responses to the viral nucleoprotein (N). The presence of anti-SARS-CoV-2 antibodies in the blood is therefore considered to be a good measure of immunosuppression for a vaccine candidate. This requires fast, reliable and sensitive methods to detect SARS-CoV-2 neutral antibodies.
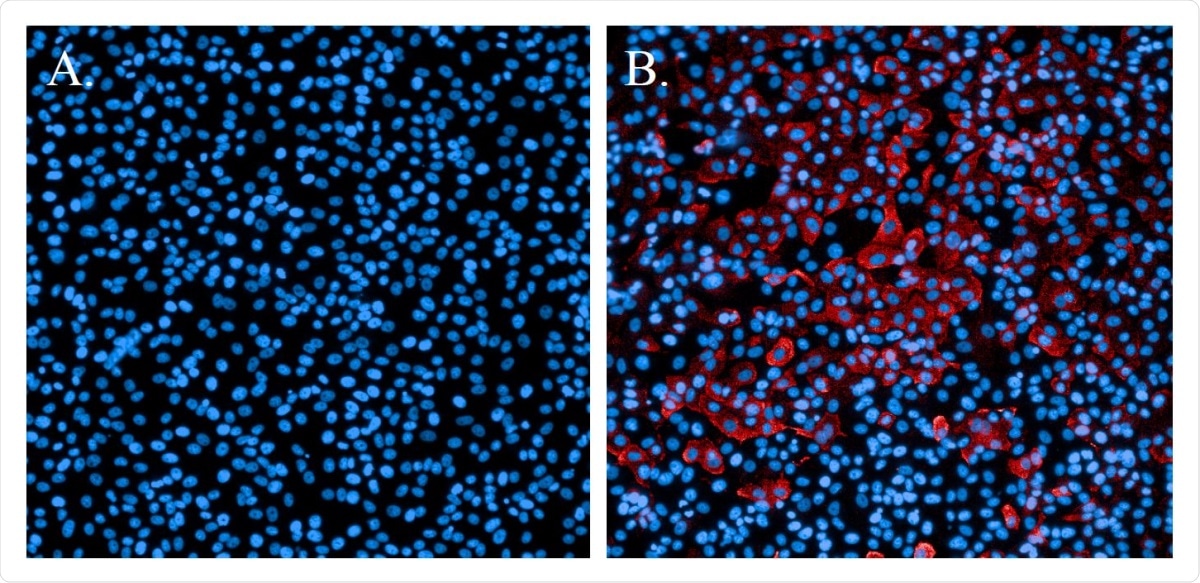
Immunofluorescence staining of SARS-CoV-2 infectious cells. A. Uninfected cells sculpted with Hoechst nuclear stain (blue). B. Cells that were SARS-CoV-2 and tested with SARS-CoV N-protein specific antibody and Alexa594 secondary antibody (red). Cells were resistant to Hoechst nuclear stain (blue).
The FRNA is a highly specific, quantitative and rigorous assay for the evaluation of neutral antibodies in a test sample. The assay measures individual cells that are initially infected with the virus and do not require several cycles of viral replication.
As an acetic assay, it does not rely on thematic diagnosis of cell cytopathic effects or development of heterogeneous or immunofoci tablets using a low number of infectious grains per source. The researchers tested immigrants in virus control and cell control and assessed the variability and specificity of the assessment.
Because the assay uses 12-step dilution schemes and four-parameter logical analysis to measure specific NT50s, it provides a more detailed understanding of neutralization potential, particularly with respect to monoclonal antibodies or nanobodies.
In this study, the researchers described the development of a SARS-CoV-2 semi-high-throughput neutralization assay that takes advantage of the capabilities of a high-content imaging system to measure the number of infectious cells in individual sources. Importantly, the researchers showed that the assay has been rapidly modified for use with a number of virus modifications. Such assessments greatly facilitate and accelerate preclinical vaccine studies and clinical trials.
Initially, during the COVID-19 emergency, the U.S. FDA (Food and Drug Administration) approved an Extended Access Program (EAP) to treat COVID-19 patients using plasma from individuals with a neutral titer of 1: 160 or less. higher. This program treated more than 94,000 patients across the U.S. Last August, the FDA issued an Emergency Use Authorization (EUA) to allow therapeutic plasma treatment of outpatient COVID-19 patients the context of clinical trials.
The researchers state that this assay has no thematic explanation and is therefore more accurate than most other wild-type virus-neutralization assays.
“While variations in live virus assessments will always be due to the nature of the biological system, using a rigorous statistical approach to inform data acceptance can be detrimental. potential effects of poor infectious efficacy, piping errors, marginal effects, and inconsistencies. staining. ”
* Important message
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical / behavioral practice, or treated as fixed information.
Magazine Reference:
- Richard S. Bennett, Elena N. Postnikova, Janie Liang, Robin Gross, Steven Mazur, Saurabh Dixit, Vladimir V. Lukin, Greg Kocher, Shuiqing Yu, Shalamar Georgia-Clark, Dawn Gerhardt, Yingyun Cai, Lindsay Marron, Michael R. Holbrook (2021) Scalable, Micro-Neutralization Evaluation for Qualitative Evaluation of Virus-Neutralizing SARS-CoV-2 (COVID-19) Antibodies in Human Clinical Samples. bioRxiv 2021.03.05.434152; doi: https://doi.org/10.1101/2021.03.05.434152, https://www.biorxiv.org/content/10.1101/2021.03.05.434152v1