
Researchers in China and Australia have reported the discovery of novel bat coronaviruses that reflect the diversity and complex evolutionary history of these viruses.
In early 2020, the novel acute coronavirus syndrome 2 (SARS-CoV-2) was identified as the causative agent of the 2019 coronavirus disease outbreak (COVID-19) that first began in Wuhan, China, at the end of December 2019.
A combination of sequencing studies and subsequent genome sampling identified several SARS-CoV-2-associated coronaviruses in wildlife species that together indicated an overestimation of phylogenetic and genomic diversity of coronaviruses.
Now, Weifeng Shi from Shandong First Medical University & Shandong Academy of Medical Sciences in Taian, China and colleagues have performed meta-transcriptomic analysis of samples collected from 23 species of bats in Yunnan district in China during 2019 and 2020.
“Our study highlights both the remarkable diversity of bat viruses at the local level and the prevalence of SARS-CoV-2 and SARS-CoV relatives in wildlife species in a wide geographical area. Southeast Asia and Southern China, ”the team says.
“These data will help guide research efforts to determine the origins of SARS-CoV-2 and other pathogenic coronaviruses. ”
A pre-printed version of the research paper can be found on the bioRxiv server, while the article is subject to peer review.
Bats are well-known hosts of coronaviruses
Guest bats are based on a wide range of viruses that are known to cause serious infections in humans, including Hendra virus, Ebola virus and, in particular, coronaviruses.
Four of the seven known human coronaviruses have a zoonotic origin. These include the SARS-CoV virus which is responsible for the 2002 to 2004 SARS revolution and the Middle East respiratory coronavirus (MERS-CoV) which is responsible for several outbreaks of severe respiratory illnesses. across the Middle East since 2012.
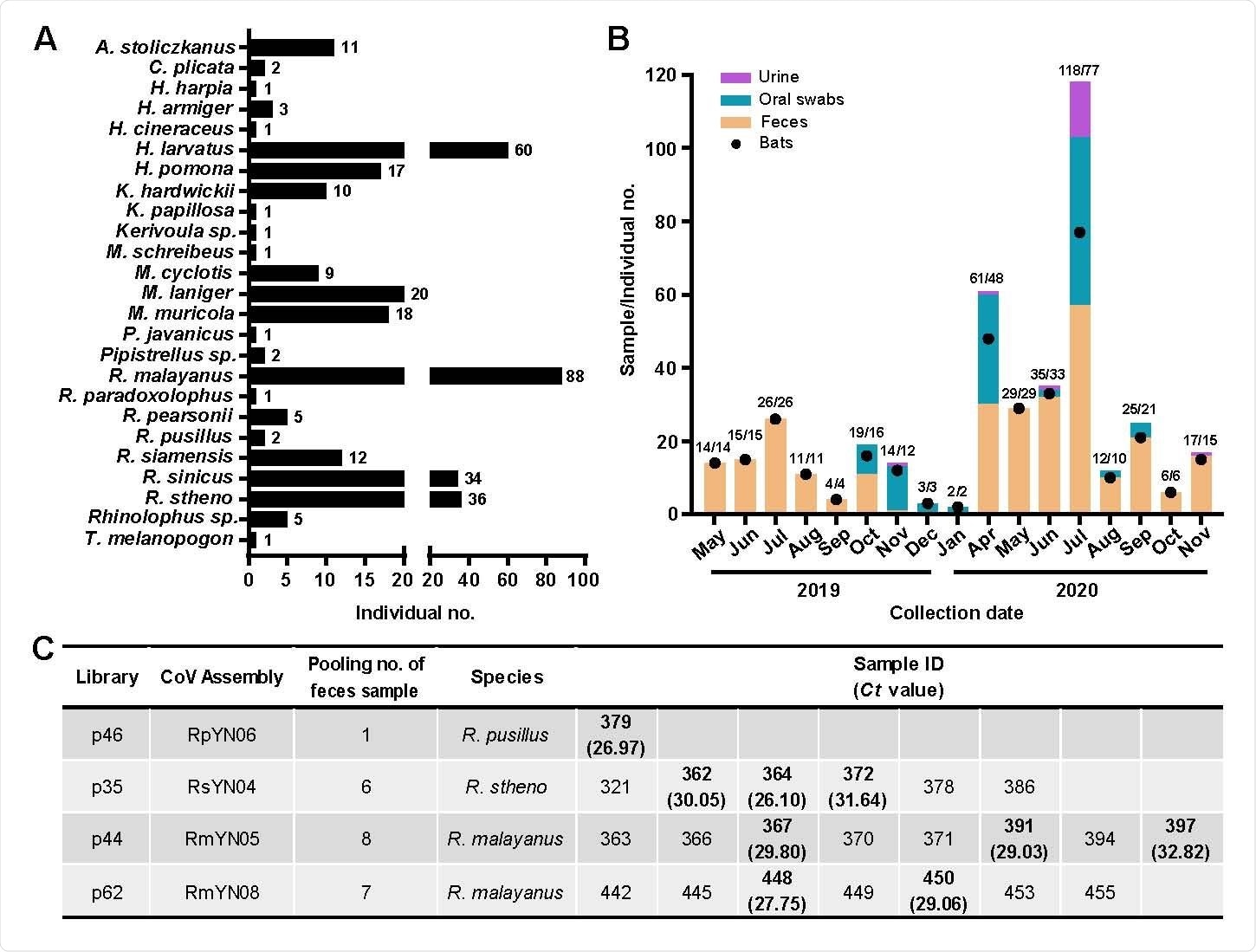
Sampling information and detection of SARS-CoV-2-like viruses in individual bat fecal samples. (A) Examples of different species of bats caught live in the Yunnan region from May 2019 to November 2020. (B) Numbers of samples collected from different periods (orange column – feces; green – oral swab; light purple – urine). Individual bat numbers are indicated by black dots and are related to the y-axis. The related numbers are in the form of individual sample numbers / bat numbers. (C) Identification of advanced samples of SARS-CoV-2 virus using qPCR.
Although bats are the most likely host of these coronaviruses, the appearance in humans has also included “intermediate” hosts such as the palm civet (in the case of SARS-CoV). and dromedary camels (MERS-CoV).
In early 2020, the novel SARS-CoV-2 virus was named the causative agent for a pneumonia outbreak in Wuhan, China, which quickly turned into a global pandemic.
A combination of sequencing studies and genome sampling identified the reversal of several SARS-CoV-2-associated coronavirus in wildlife species.
These included the RaTG13 virus found in the Rhinolophus affinis bat, the most closely known most closely related component of SARS-CoV-2 throughout the viral genome as a whole.

Ecological modeling of the geographical distribution of 49 species of Rhinolophid bats. (A) Models of 49 Rhinolophus bat species predicting diversity in five regions covering mainland Southeast Asia, the Philippines, Java-Sumatra, Borneo and Sulawesi-Moluccas. The color of the map represents the richness of species, with up to 23 species expected. (BF) Placemate (B) host species RaTG13 R. affinis, (C) host species RpYN06 R. pusillus, (D) host species RmYN02 R. malayanus, (E) host species RacCS203 R. accuminatus, and (F) the host sex STT182 and STT200 R. shameli. The yellow area represents the expected range of each species.
Viruses associated with SARS-CoV-2 have also been identified in several other Rhinolophid bats, including R. shameli bats sampled in Cambodia, R. cornutus bats sampled in Japan, and R. acuminatus bats sampled in Thailand.
“Together, these studies show that bats over a wide range of Asian harbor coronaviruses are closely related to SARS-CoV-2 and that the phylogenetic and genomic diversity of these viruses appears to have been implicated. reduce ”said Shi and team.
What did the researchers do?
To further study the diversity, ecology, and evolution of bat viruses, Shi and colleagues performed a meta-transcriptomic study of 411 samples collected from 23 bat species in a small area (111100 hectares) side. within the Yunnan region of China, between May 2019 and November 2020.
The team identified coronavirus contigs (sets of overlapping DNA sequences) in 40 of 100 sequence libraries, including seven libraries with contigs that could be mapped to SARS-CoV-2.
The team found modern coronavirus genomes
From that data, the researchers collected 24 full-length novel coronavirus genomes, including four SARS-CoV-2-related genes and three SARS-CoV-related genes.
Notably, one of these novel bat coronaviruses – RpYN06 – exhibited a 94.5% order identity to SARS-CoV-2 throughout the entire genome. Furthermore, in some individual genes (ORF1ab, ORF7a, ORF8, N, and ORF10), RpYN06 was the closest relationship of SARS-CoV-2 identified to date.
However, at the genomic scale, low order identity in the RpYN06 spike gene produced the second closest relative of SARS-CoV-2, alongside RaTG13. The spike protein is the main structure used by SARS-CoV-2 to bind to and influence host cells.
Differences in the overall order of the spike gene
The researchers say that while several SARS-CoV-2-like viruses have previously been identified in different wildlife species, none of them are very similar (> 95%) in full sequence identity. the spike gene.
In fact, while the other three SARS-CoV-2-related viruses identified here were almost identical in sequence, the spike protein sequences formed an independent line separated from sarbecoviruses. known by a relatively long branch. Sarbecovirus is a viral subgenus or coronaviruses containing SARS-CoV-2 and SARS-CoV.
“Together, these results highlight the very high and predisposed genetic diversity of the sarbecovirus spike proteins, which tend to show the variable flexibility,” Shi and her colleagues wrote. .
The researchers say studies have previously shown that host mutation of coronaviruses among bats often occurs.
“Catching that multi-virus bat species can increase the difficulty in detecting the origin of SARS-CoV-2 and other pathogenic coronaviruses,” they add.
What else did the study find?
Ecological modeling predicted that there would be up to 23 species of Rhinolophus bats in much of southeast Asia and southern China, with the largest sites for high bat diversity extending from South Lao and Vietnam into southern China.
The researchers say that, in addition to Rhinolophids, this wide geographical area in Asia is rich in many other bat families and other wildlife species that have been shown to be susceptible to SARS-CoV. -2 in vitro.
“It is therefore essential that further research efforts cover a wider range of wildlife in this region to help monitor the ongoing outbreak of SARS-CoV-2, SARS-CoV and other pathogenic viruses from animals to humans, ”they conclude.
* Important message
bioRxiv publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical practice / behavior, or be treated as information established.