COVID-19 infection is caused by the novel true coronavirus acute coronavirus syndrome 2 (SARS-CoV-2). COVID-19 was declared a pandemic by the World Health Organization in March 2020. As of today, more than 106 million SARS-CoV-2 infections and more than 2.31 million deaths due to disease have been reported. COVID-19 worldwide.
The chronic pandemic COVID-19 emphasizes the importance of molecular analysis to understand the evolution of the virus and to monitor and plan epidemiological responses.
An epidemiological approach to study aims to determine the origin of revolution and the nature of the transmission chains. Some tools that are particularly suitable for this purpose are phylogenetic trees, maps and timelines. However, a pathogen-related approach that focuses on vaccines, treatments, and sprays requires method-specific imaging and offers a number of specific applications. Public health research organizations or researchers from universities often study the same pathogens but may need slightly different images, for example, to study different genes, which may require time adjustments. For this reason, easy visibility, quick inspection, and useful filtering of the latest series are essential for this approach.
Making series information more accessible to epidemiologists and virologists
Recently bioRxiv* preprint research paper, German scientists discuss how they made series information easier for epidemiologists and virologists. The researchers, working with the German public health authorities, developed a molecular screening device called CovRadar for SARS-CoV-2 spike proteins. The tool has a checklist and a web application.
According to the researchers, CovRadar is very flexible and can handle a lot of data, and users can customize it according to their needs. Both the pipeline and the app can be installed and run on local systems so that streams with accessible access can be imported, and all functions are offered at covradar.net.
The coronary virus spike protein has a receptor binding domain (RBD), which is a target for several vaccine candidates. CovRadar enables more than 250,000 series to be viewed and viewed.
It first identifies the areas of interest using local alignment and then develops multi-layered alignment, including variations, consensus sequences, and phylogenetic trees and then display the results in an interactive app, making a useful and flexible report.
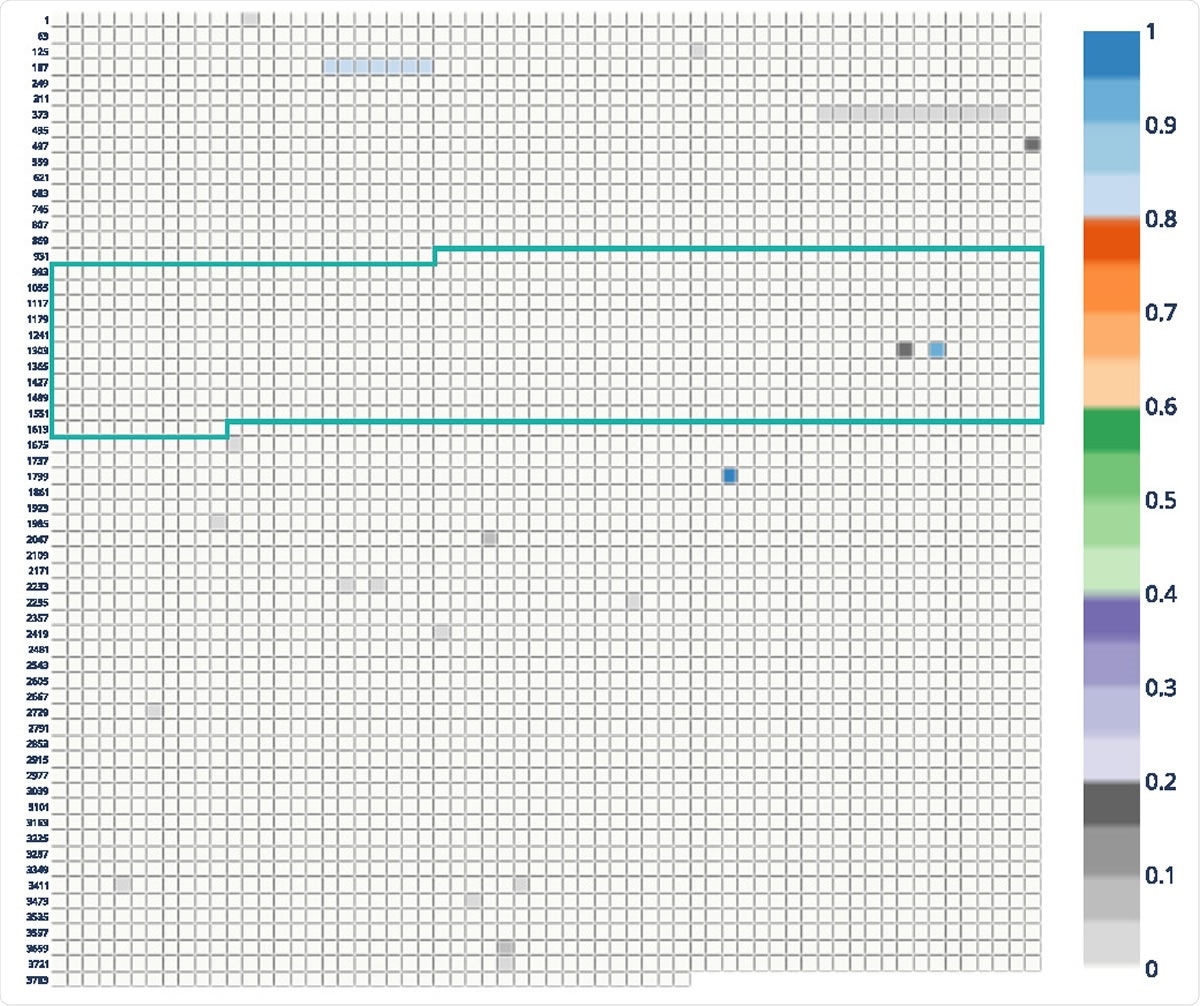
Allele frequency plot of Danish mink lines with frame-bound receptor attachment (RBD). Each block represents a nucleotide in the MSA of spike gene sequences. Coordinates on the left are related to the position of MSA. The plot shows frequencies> 0: 8 for nucleotide positioning according to codes 68-70, 453 and 614.
CovRadar has a PDF-like reporting format with interactive capabilities
The interactive report on SARS-CoV-2 spike protein is an application written in Flask and Dash and is linked to the MySQL database. The web app layout is similar to a PDF report with interactive capabilities, including filtering, printing, and navigation in the side menu bars. Users can save the tables and plots in excel and PNG page format, respectively, for use in other ways. All elements, including plots and boards, are directly editable.
CovRadar can help with situations where zoonotic movements with specific host mutations are possible. A number of mutations are currently being considered to determine whether they are related to mink and their importance for the vaccines in development. With the help of CovRadar, the researchers analyzed the conditions in typical Danish mink data with high allele frequencies and compared the data with human data both before and after the revolution. mink. They found an increase of other alleles mainly at codon 453 and 68-70 positions, which may be related to Y435F and del69-70.
CovRadar is available for free at https://covradar.net. The open source code is available at https://gitlab.com/dacs-hpi/covradar.
“CovRadar was designed to help with molecular tracking of Corona spike proteins that have been used as a target for most vaccine candidates. ”
* Important message
bioRxiv publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical practice / behavior, or be treated as information established.