Using more than 100,000 virus genome sequences uploaded to the GISAID database, researchers found approximately 9,000 mutations in the RBD protein spike virus.
Among the coronaviruses known to infect humans, some affect the upper respiratory tract, and some lower respiratory tract causes moderate to severe infections. The acute respiratory coronavirus 2 (SARS-CoV-2) syndrome that is responsible for the COVID-19 pandemic is an infectious virus that usually affects the lower respiratory tract. which leads to mild symptoms such as sore throat to severe infection, and even death.
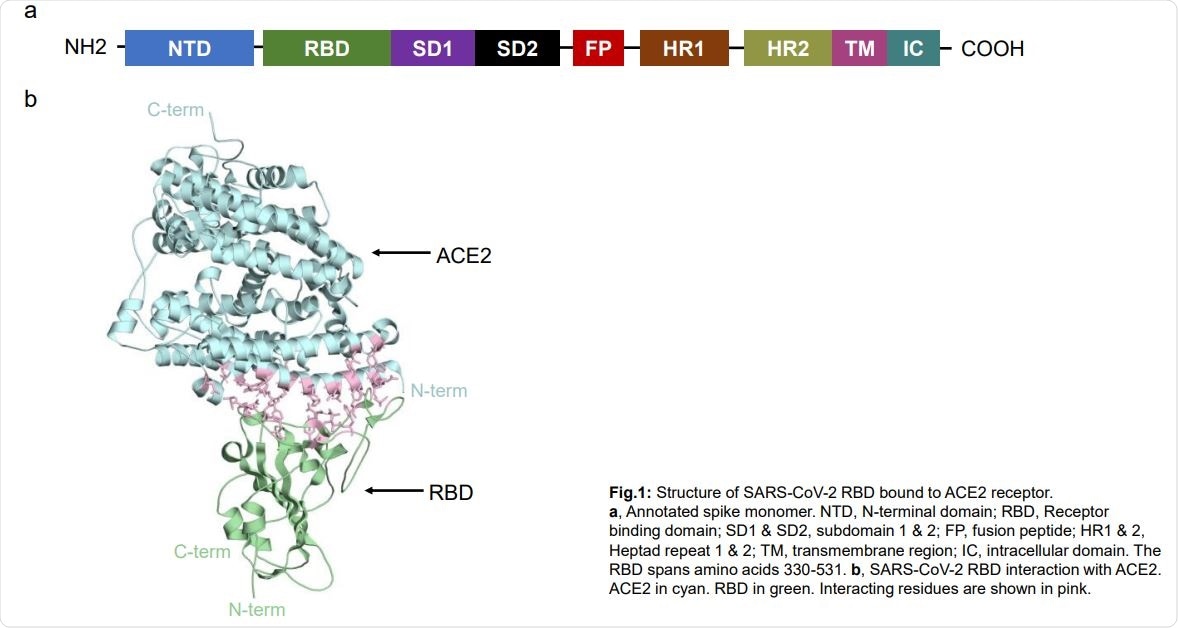
The virus’s spike proteins interact with host receptors such as the angiotensin converting enzyme 2 (ACE2) and TMPRSS2 protease, which allow the virus to bind and then dissolve with the host cell. The virus releases RNA into the host cytoplasm, where it reproduces.
The spike protein has two subunits, S1 containing the receptor binding domain (RBD), which interacts with ACE2, and S2. Variations in the RBD or ACE2 can affect virus infection and disease status.
To understand RBD mutations and their impact on antibody interactions, a team of researchers from the University of Toronto analyzed the RBD virus using the SARS-CoV-2 genome information in the Enterprise database. Global has been sharing all flu data (GISAID). They report their findings in a research paper posted to bioRxiv * preprint server.
RBD mutations
The researchers found more than 110,000 RBD sequences uploaded to the database as of October 15, 2020. After removing duplicates, gaps with gaps, and other inappropriate sequences, the team found 106,941 sequences. The nucleotide sequences were aligned with the RBD of the reference constraint and analyzed.
Compared to the Wuhan reference strain, the team detected 9,275 mutations on the RBD, resulting in 9,064 SARS-CoV-2 mutants. Approximately 75% of the nucleotide mutations were at G1430A, 6.8% were C1317A, and 0.9% was G1144T. The most common mutation was a change from G to A, occurring in about 7% of the mutants, followed by changes from C to A, then G to T, and C to U.
Of all nucleotide mutations, about 98.3% altered the amino acid sequence of the RBD. The rest of the mutations were meaningless. The G1430A mutation resulted in the mutant S477N, representing 76.5% of all amino acid mutations. The C1317A and G1144T mutations resulted in modifications of N439K and A520S, respectively.
SARS-CoV-2 RBD structure attached to ACE2 receptor. a, Monomer spike with notes. NTD, N-terminal domain; RBD, Receptor binding domain; SD1 & SD2, subdomain 1 & 2; FP, peptide fusion; HR1 & 2, Heptad 1 & 2 repetition; TM, transmembrane area; IC, intracellular domain. The RBD span amino acid 330-531. b, SARS-CoV-2 RBD interaction with ACE2. ACE2 in cyan. RBD in green. Interactive remains are shown in pink.
Previous studies have shown that the S477N and N439K mutants show greater affinity for ACE2. The analysis showed that these mutations represent 6.7% and 0.6% of the total genomes in the database, respectively, with a 70-fold increase in the presence of S477N mutation. This indicates that selection may play a role in RBD mutations. In total, the authors found 13 mutations that led to an increase in binding affinity with ACE2.
The S477N mutation has been detected in 14 countries worldwide, with approximately 69% of Australia’s series with this simulation. N439K was seen in 10 countries, with the highest occurrence in England. They did not find both mutations occurring on the same genome. The team also found that all S477N or N439K mutations also had a D614G mutation in the spike protein.
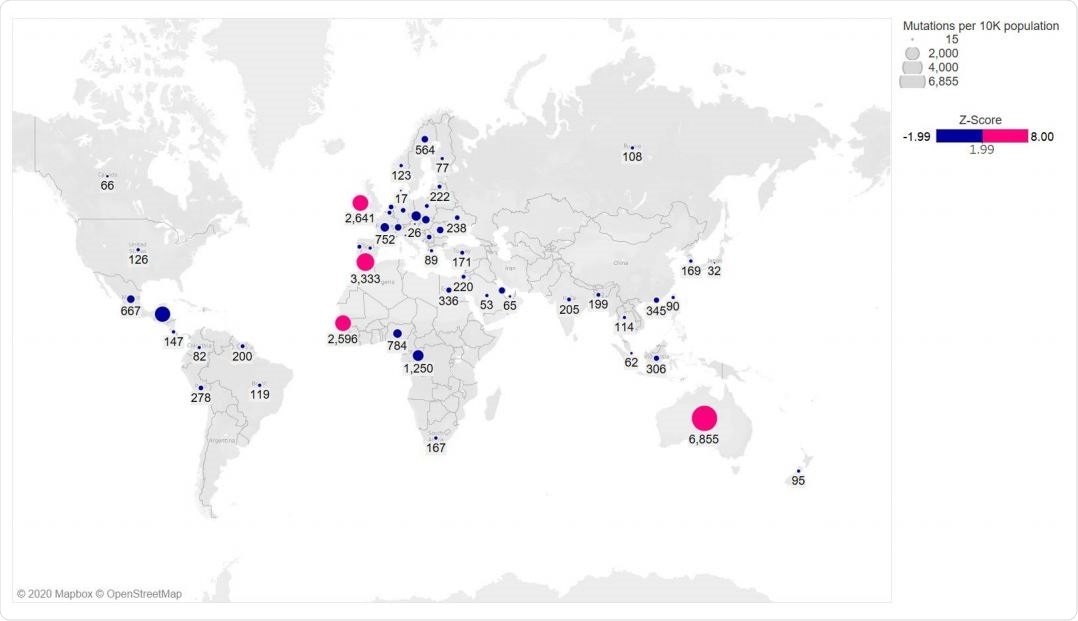
Global distribution of RBD mutations
No mutations at residues that increase binding to ACE2
What surprised the authors was that they did not find any mutations at the Q498 residue of the RBD because Q498H, Q498Y, and Q498F all increase binding affinity to ACE2. They also did not detect or detect in numbers now other mutations, such as N501F, Y453F, T385R, Q493M, and Q414A, which increase RBD binding to ACE2.
This is probably because mutations at Q498 would be unbalanced in virus fitness and could reduce virus transmission according to the spread-death trade theory. If this were correct, the SARS-CoV-2 mortality rate would decrease over time, and the spread would increase.
When the team analyzed human antibody interaction sites with the RBD, they found that 75% of the interaction sites of 10 human antibodies had mutations. Their analysis also showed that the antibodies worked with widespread transmission of the RBD. None of the mutations had a direct interaction with every antibody they studied.
“This suggests that vaccination with wild-type and possibly any RBD point mutant that preserves structure is likely to develop RBD antibodies that are sufficient to bind and dissolve disease,” the authors wrote. However, it remains to be seen how the mutations will affect virus infection and transmission.
* Important message
bioRxiv publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical practice / behavior, or be treated as information established.