.jpg)
The genomic sequence of coronavirus respiratory syndrome 2 (SARS-CoV-2) has helped to monitor movements in viral isolates across the globe.
With the advent of the D614G mutation, the SARS-COV-2 G-strain became transferable. Studies show that this variant has increased cell binding and viral transmission within in vitro models. More specific strains of SARS – CoV – 2 with further changes have emerged in recent months, spreading rapidly in the United Kingdom.
These different variants are in the spike (S) gene, specifically N501Y. The rapid onset and abrupt changes in the S gene have raised concerns about changes in the pattern of the disease and the potential difference in how COVID- patients become. 19 dealing with vaccines or antibody treatments.
Genomic analysis of SARS – CoV – 2 in Columbus, Ohio, reveals a parallel shift in coverage 20C> 20G with 3 new variations
Researchers from the United States recently reported the results of a SARS – CoV – 2 genomic study conducted between April 2020 and January 2021 in Columbus, Ohio. Their work is released as an introduction to the bioRxiv* server.
The study data show a recent parallel shift in the largest 20C> 20G shoreline with 3 new changes. It also shows that the novel viral presence is separated by the S version N501Y but differs from the RA-B.1.1.7 (20I / 501Y.V1) and the South African variant (20H / 501Y.V2).
“The same N501Y mutation was subsequently detected in a 20C clade strain in South Africa, where it was associated with a different set of additional S modifications, with the Next Strain designation as 20H / 501Y .V3. “
One of these isolates has become the largest virus detected in nasopharyngeal swabs between December 2020 and January 2021 and contains S p.Gln677His, membrane glycoprotein (M) p.Ala85Ser (Q677H) , and nucleocapsid (N) p.Asp377Tyr (D377Y). The other solitary has S N501Y and ORF8 Arg52Ile (R52I), the two markers of UK snoring – B.1.1.7 but not all other viral mutations. It shares several mutations with the 20C / G clade viruses present in Ohio by December 2020. Both of these viruses currently appearing in the U.S. contribute to the proliferation of Spike gene mutations throughout worldwide and supports a number of independent builds of S N501Y during this deployment phase.
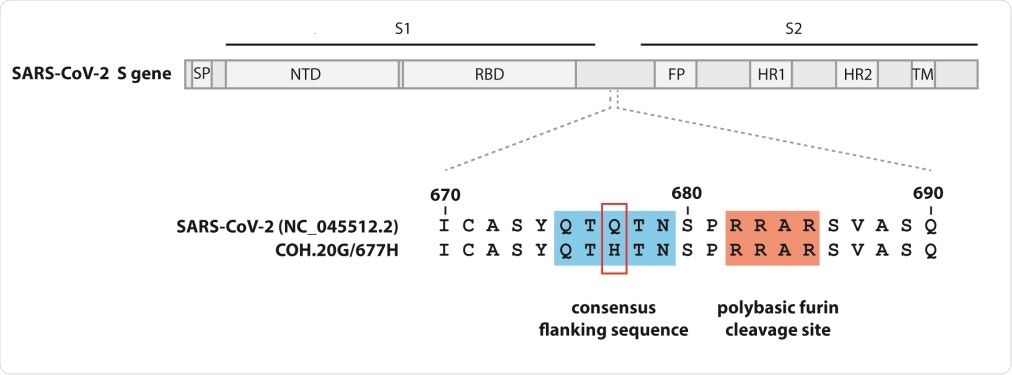
Q677H mutation site in Spike gene in QTQTN motif conservation / furin clearance site. Signal peptide (SP), N-terminal domain (NTD), receptor-binding domain (RBD), peptide fusion (FP), heptad 1 replication (HR1), heptad 2 replication (HR2), and transmembrane domain (TM).
The rapid release of Asp377Tyr N modification deserves closer attention
The researchers report a distinct viral presence from December 2020 in Columbus, Ohio that has acquired the S version N501Y. This amino acid mutation was first detected in a UK clinical sample along with other novel spike modifications and a 20B clade spine. The mix was named as B.1.1.7 strain and NextStrain specification as 20I / 501Y.V1. The same N501Y mutation was subsequently detected in a South African 20C clade strain associated with a different set of additional S modifications.
At the end of December, we found one sample with a 20G strain backbone that had undergone S N501Y and ORF8 R52I mutations (named COH.20G / 501Y, Figure 1, arrow), both of which are present in the UK – B.1.1. 7 strain. “
The frequency of these two variables started to rise sharply at the end of December 2020 and the S 501Y mutation was thought to be the reason behind further spreads. The virus has a 20G backbone with the imitation S N501Y found in Columbus (COH.20G / 501Y) but does not contain most of the consensual changes reported in 20I / 501Y.V1 and 20H / 501Y.V2.
They also report the emergence of the SARS – CoV – 2 virus variant with a predominant 20C / G clade and single mutations in the S, M, and N genes in Columbus, Ohio, in December 2020. A mutation of N677H affecting the QTQTN series sequence adjacent to a polybasic fur coat site crossing the S1 and S2 junction. It has rarely been reported in NextStrain and in other publications. Studies have reported the elimination of a QTQTN motif that can affect viral properties. According to the authors, the Asp377Tyr N modification has not been reported much before, and its rapid emergence deserves closer attention.
“We also report that the SARS – CoV – 2 virus strain was predominantly derived by a 20C / G clade spider that has single mutations in the S, M and N genes (Gln677His, p.Ala85Ser, and p. Asp377Tyr, respectively) in Columbus, Ohio in December. “
* Important message
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical / behavioral practice, or treated as fixed information.
Magazine Reference:
- Specific patterns of exposure of SARS-CoV-2 Spike variants including N501Y in Clinical Samples in Ohio Columbus, Huolin Tu, Matthew R Avenarius, Laura Kubatko, Matthew Hunt, Xiaokang Pan, Peng Ru, Jason Garee, Keelie Thomas, Peter Mohler, Preeti Pancholi, Dan Jones bioRxiv 2021.01.12.426407; doi: https://doi.org/10.1101/2021.01.12.426407, https://www.biorxiv.org/content/10.1101/2021.01.12.426407v1