.jpg)
The severity is the result of severe respiratory coronavirus 2 (SARS-CoV-2) infection, with the majority of asymptomatic individuals and others experiencing severe influenza and multiple organ failure. Hospitality genetics plays a role in disease progression and prognosis. A paper was recently uploaded to the preprint server medRxiv* with Dubé et al. (1st March 2021) identifies several alleles associated with the natriuretic peptide receptor C (NPR3) receptor that may affect disease severity.
How was the study conducted?
The group worked with 4,488 outpatients with high COVID-19 risk over 40 years of age and with at least one other comorbidity such as diabetes or obesity. The group was divided into two equal doses, one of which received a placebo, and the other received 0.5 mg of colchicine, an anti-inflammatory, daily for a month. Patients were assessed for symptoms of disease 15 and 30 days into the trial.
The same patients were asked to participate in an additional study to help identify genetic determinants in COVID-19 results. Genetic data were collected from those who gave consent and a COVID-19 result was found for each individual. The database was further updated by removing genetically related samples or those without positive COVID-19 PCR tests, leaving 1,855 for analysis.
78% of the total participants reported being free of COVID-19 symptoms in the 30-day period, with the authors noting that, on average, women experienced symptoms longer than men. . Interestingly, 0.3% and 5.1% of colchicine test participants died or were hospitalized, respectively, although only 3.1% of those who went on to be admitted to the genetic study were hospitalized , and none died. Unfortunately, as the authors point out, healthy partner bias means that less healthy patients are less likely to participate in the additional genetic study.
The time for postoperative symptom relief was recorded among the genetic analysis group, and in-silico methods were used to identify two common genetic differences associated with shorter disease duration.
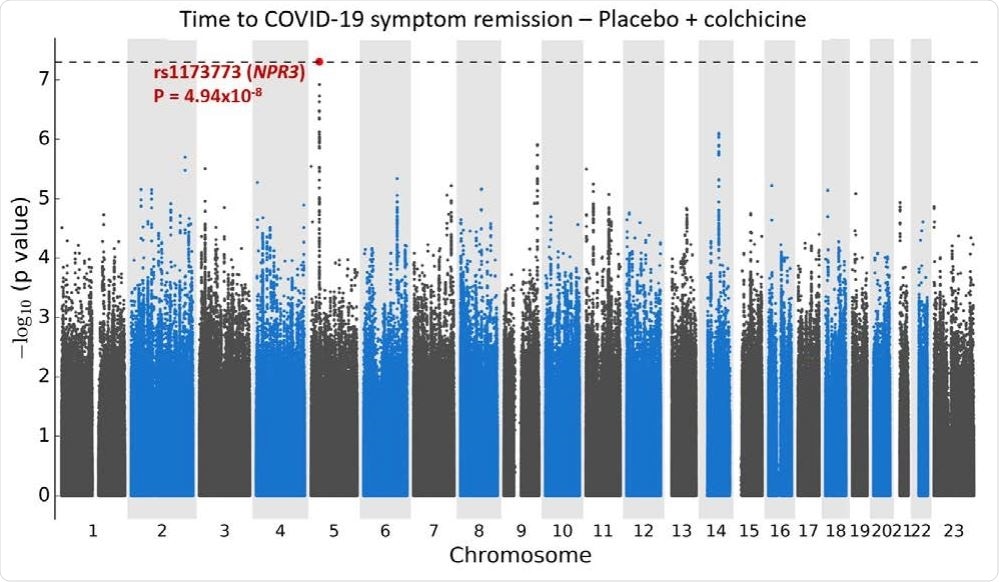
Manhattan plugs for the GWAS time to relieve COVID-19 symptoms. a. Using Cox proportional risk retrieval with 1723 subjects from colchicine and placebo arms COLCORONA study, controls for study arm, gender, age, and 10 key components for genetic ancestry, with 6,392,715 genetic differences of min. -type frequency ≥ 5%.
What alleles have been identified?
Two loci of interest were identified by the group, located at 5p13.3 and 9q33.1. The chromosome 5 variant spans the NPR3 gene, which is involved in the enlarged renin-angiotensin system. Differences in NPR3 have also been associated with height, blood pressure, and the critical mass of a person’s lungs. Those with copies of the small allele had a shorter release time than those with the main allele, with 44%, 54% and 59% of patients becoming healthy 15 days after infection if 0 , 1, or 2 copies of their allele, respectively.
The chromosome 9 allele has also been associated with a person’s height and is a risk factor for venous dysfunction. In this case, the locus difference had only a visible effect on symptom relief in the non-placebo group, which the authors suggest may be due to colchicine administration. Several similar inflammatory pathways are activated by both downstream effects of chromosome 9 and colchicine, which may exacerbate the genetic effect in the recipient group. drug. Within the placebo group, 46%, 62% and 64% of patients had symptom relief within 15 days when the individual had 0, 1, or 2 copies of the allele .
Since the length of the signal is a strong predictor of disease severity, this newly discovered genetic risk factor may help predict outcome. Deeper classification of a person’s risk factors by genetic analysis will allow future treatments to be designed, but in this particular case more follow-up studies with more partners are needed to confirm the findings. .
* Important message
medRxiv publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical practice / behavior, or be treated as information established.
Magazine Reference:
- Genetic relief of symptoms in outpatients with COVID-19, Marie-Pierre Dube, Audrey Lemacon, Amina Barhdadi, Louis-Philippe Lemieux Perreault, Essaid Oussaid, Geraldine Asselin, Sylvie Provost, Maxine Sun, Johanna Sandoval, Marc-Andre Legault , Ian Mongrain, Anick Dubois, Diane Valois, Emma Dedelis, Jennifer Lousky, Julie Choi, Elisabeth Goulet, Christiane Savard, Lea-Mei Chicoine, Marieve Cossette, Malorie Chabot-Blanchet, Marie-Claude Guertin, Simon de Denus, Nadia Bouabdallaoui, Richard Marchand, Zohar Bassevitch, Anna Nozza, Daniel Gaudet, Philippe L L’Allier, Julie Hussin, Guy Boivin, David Busseuil, Jean-Claude Tardif, medRxiv, 2021.02.24.21252396; doi: https://doi.org/10.1101/2021.02.24.21252396, https://www.medrxiv.org/content/10.1101/2021.02.24.21252396v1