Chronic coronavirus disease 2019 (COVID-19), caused by the severe respiratory coronavirus 2 (SARS-CoV-2) syndrome, continues to spread worldwide. The release is prompted by the appearance of newer, more infectious changes such as the B.1.1.7 version, which came out in the UK towards the latter part of 2020.
A new introduction to the medRxiv * server outlines the upcoming rise in leadership of the B.1.1.7 variable in the US. Understanding this development will help shape protective action to reduce infection and mortality rates.
The variable B.1.1.7
The SARS-CoV-2 Variant of Concern (VOC) line 202012/01 (aka 501Y.V1; B.1.1.7) was first identified in the second half of 2020. It was detected early due to large series and rapid release of SARS-CoV-2 in the UK, as part of that country’s genomic analysis program.
The UK variant shares the N501Y mutation with the mutation in South Africa and Brazil, but there are additional mutation as well. These ‘signature’ mutations include delete at 69/70, delete at 144, along with A570D, D614G, P681H, T716I, S982A, and D1118H.
The RA variant is 40-70% more infectious than the other variants, possibly because the N501Y strengthens the binding affinity of the spike protein SARS-CoV-2 with the host cell receptor, enzyme 2 converted angiotensin (ACE2) . In addition, it can cause up to 30% higher mortality rates.
The polymerase chain reaction (PCR) experiments used for the detection of SARS-CoV-2 infection could provide an important indication of the presence of such viral sequences. This is because the target probe sites used in the test have very different sequences. Elimination of the RA variant is at the 69/70 spot, which usually results in undetectable on the S gene with PCR tests based on the parent sequences.
This is called S gene target failure (SGTF). In samples showing SGTF in the UK, the percentage of B.1.1.7 sequences increased from 3% to over 90%, in the period beginning the week of October 12, 2020, and ending week 30 November, 2020.
Increase in the proportion of B.1.1.7 within SGTF
The current study examined the distribution and frequency of this change in the US. The researchers selected only SGTF samples for ordering, since such samples are automatically enriched for B.1.1.7. The database contained all SARS-CoV-2-positive samples tested at Helix facilities from July 2020 onwards, approximately half a million in total.
Helix testing for COVID-19 tests for three target antigens, the spike (S), the nucleocapsid (N), and the ORF1ab (1ab open reading frame). If a sample found both the N and ORF1ab but not the spike, it is called the SGTF sample.
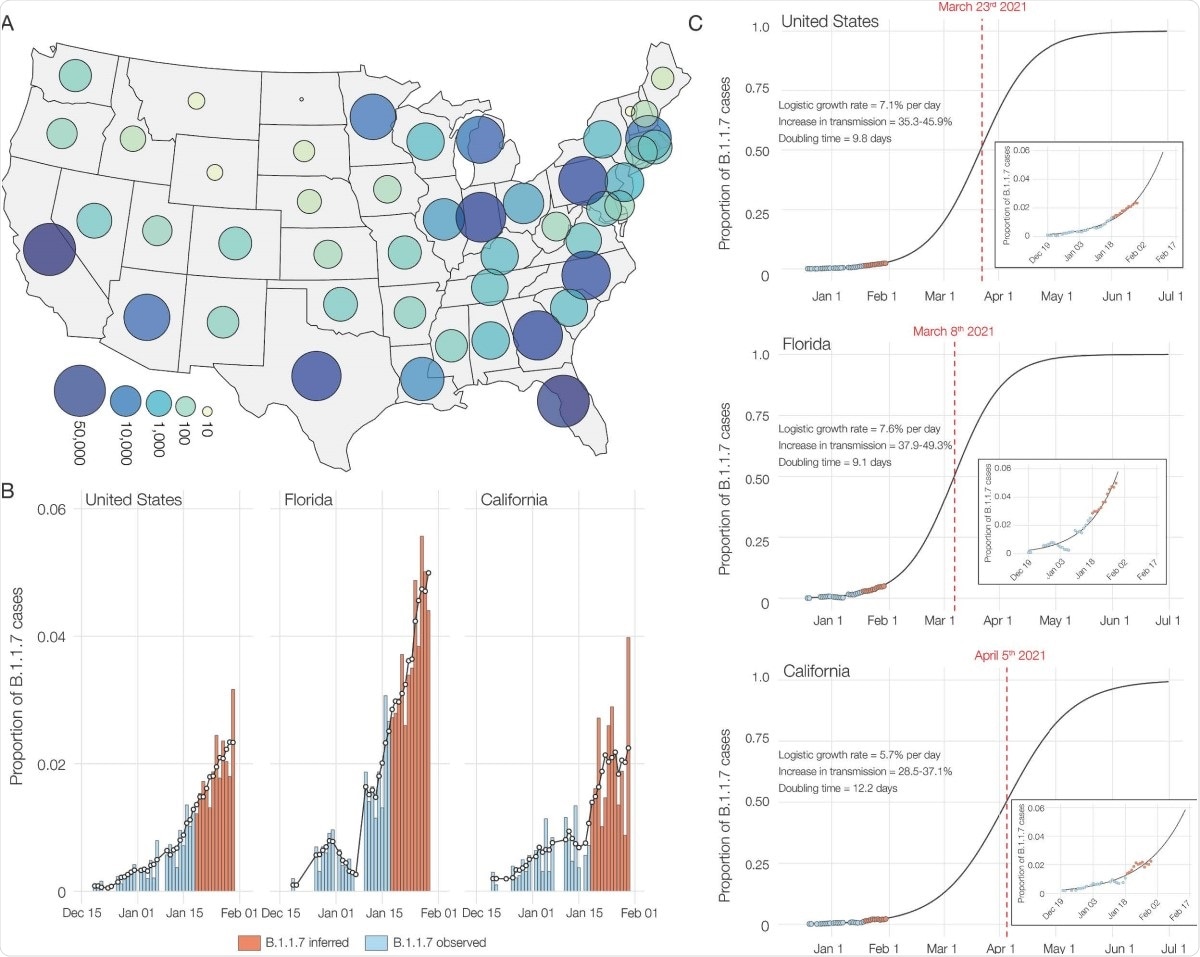
From early October 2020, SGTF began to see a consistent low frequency, comprising 0.2% of daily positive tests by October 18. In January 2021, it rose from 0.8% to 4.2% from the first week to the last one. Of all SGTF samples, 3.6% were of B.1.17 line.
Also, SGTF samples were found in 22 of the 53 states and districts in the U.S. in January 2021, accounting for more than 1% in many areas, but with uneven circulation.
Mark B.1.1.7 with series
Following the 460 SGTF samples obtained from December 2020 through January 2021, they found that 45% of the B.1.1.7 variance was across ten states. This difference has been reported to be caused by at least one case in 33 states and districts, in total, using data from all test laboratories.
In the current study, nearly half came from California, and a slightly smaller number from Florida. These two states accounted for nearly 180 of the B.1.1.7 sequences.
Typically in the Florida series there was a K1191N mutation in addition to, in one case, a Q493K waterproof escape mutation.
Higher growth rate of B.1.1.7 in the US
The researchers also found that B.1.1.7 accounted for 90% of all SGTF samples by mid-January 2020, from a high of 95% in California to a low of 70% in Florida. By the end of the month, this variant caused 2% of all COVID-19 cases in the country, with an increase in transmission of 35-46%. The doubling time for this variable was estimated to be 1010 days in the U.S. as a whole, 1212 days and 99 days in California and Florida, respectively.
Numerous introduction
The researchers also found that most of the B.1.1.7 series in the U.S. were in two major trenches, independently imported into California. Smaller clades represented eight other introductions, such as the GA cover, which contained three layers from Georgia. 19 single series were caused by yet another independent introduction.
The earliest introduction was revealed by the CA1 shoreline, which began the spread of a stable community in California in November 2020. Since then, several burrows have been reintroduced into the country to this time. The B.1.1.7 variant has also been spreading between states and regions since December 2020, at least.
What is the impact?
Although B.1.1.7 changes are now at a low level of the viral variables circulating in the US, it is growing at a much higher rate (an increase of 35-45%) compared to other series. . In fact, its doubling time indicates that the number of diseases caused by this variable doubles every 10 days.
These findings are consistent with those from other countries, and with the current trend of B.1.1.7 in the US, it is almost certain to become the SARS-CoV- mainline. By March 2, 2021 across many U.S. states. ”
More research, such as a robust national screening program incorporating genomic sequences, will be needed to understand the true frequency of this change in the country. It is not yet known how it was introduced, distribution instructions and originality. However, imports appear to be related to high travel times, with over a million passengers crossing checkpoints each day.
These include the main Thanksgiving time as well as the Christmas and New Year holiday periods. So it would seem that the origin came through international travel, which quickly spread in the community.
The B.1.1.7 variant now makes up 90% of SGTF samples and the increased growth rate makes it likely to soon be spread with the use of percentages. SGTF. However, greater genomic sensitivity is critical to evaluating and tracking emerging new changes, possibly with higher and ever-increasing transmission.
Unless public health action is determined and promptly, the higher release rate of these strains and the higher effective reproduction number of SARS-CoV-2 are likely to have a devastating effect on COVID-19 mortality and morbidity in the USA in a few months, if so decisive action will not be taken immediately. ”
* Important message
medRxiv publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical / behavioral practice, or be treated as information established.