In late December 2019, the 2019 coronavirus infection (COVID-19) was first detected in Wuhan City, China, causing an easily transmissible respiratory illness. By March 2020, the World Health Organization (WHO) declared their disease a global pandemic.
Caused by the acute respiratory coronavirus 2 (SARS-CoV-2) syndrome, COVID-19 has captured more than 107.48 million people worldwide. To date, more than 2.35 million have already died.
As the pandemic has progressed, the virus has evolved into mutations that are more easily transmitted and can cause more severe symptoms.
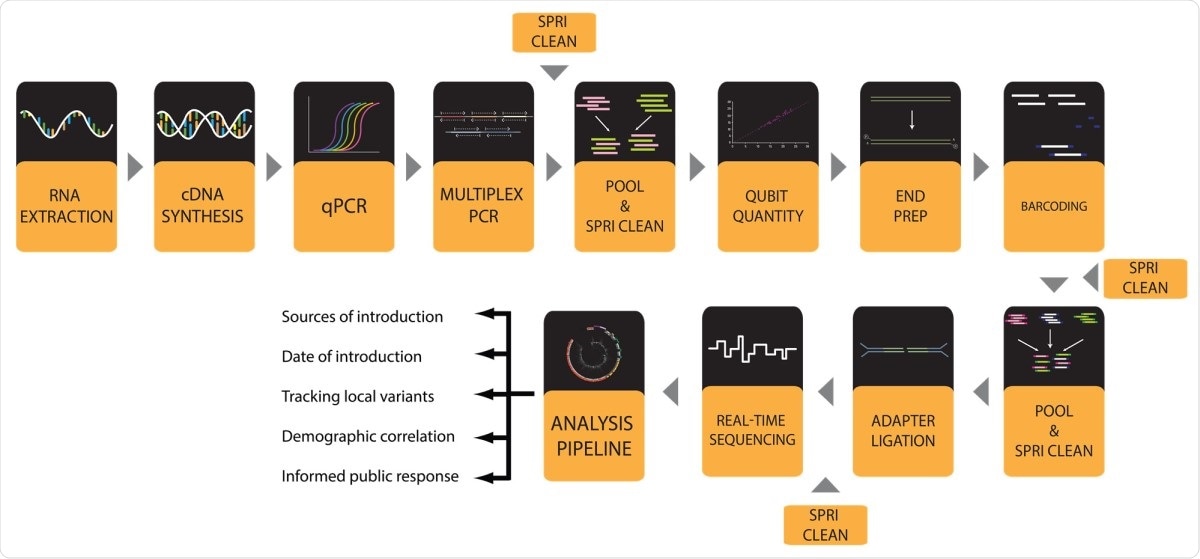
Workflow for SARS-CoV-2 viral genome sequence with MinION instrument.
Now, researchers at the University of Southern Illinois, USA, have proposed a broad set of working protocols for sequencing high-quality SARS-CoV-2 genomes in high transmission from patient samples. This can help in making sequences of the viral genome, which will help to detect mutations of the virus and help governments to implement measures to stop the pandemic.
Viral genome sequence
A sequencing of a viral genome is essential to help deal with any problems that may arise with new mutations of the virus. Viruses can develop rapidly, which can lead to higher mobility or density and pose a threat to humans around the world.
The SARS-CoV-2 has two new strains that are more potent and can cause more severe symptoms. The UK variant has already spread to other countries, causing skyrocketing cases in places like the US, while the South African variant could avoid the antibodies produced as a result of vaccination.
Thus, sequences of a viral genome can define reserved and mobile regions that may provide valuable insights into the development of effective vaccines and therapies for SARS-CoV-2.
Currently, there are many approaches to sequencing SARS-CoV-2 genomes, such as metagenomic sequencing, PCR-based proliferation techniques, and targeted enrichment sequencing. Two of these methods, particularly the metagenomic and target genomes, are expensive for generating large genomes on a large scale.
At the same time, PCR-based expansion may be a potential solution to address the limitations of alternatives. The PCR-based extension method simultaneously raises the interest target.
The study
In the present study, published on the bioRxiv * preprint server, the researchers introduced a method to classify high-quality SARS-CoV-2 genomes above from patient samples. The protocols of the research team adopted and modified the protocols, which were developed by the ARTIC Network to sequence SARS-CoV-2 viral genomes using the MiniION nanopore sequence platform from Oxford Nanopore Technologies (ONT) .
From there, the team provided a complete set of tests, including instructions on how to take every step from ribonucleic acid (RNA) extraction to data visualization. The team tested the new protocols through successfully processing more than 1,000 samples and ordering more than 600 high-quality SARS-CoV-2 genomes from Illinois.
To reach the conclusions of the study, the researchers performed the entire workflow to produce SARS-CoV-2 genomes in 96-well plate form. It begins with the extraction of RNA from viral transport media (VTM) containing nasopharyngeal swabs (NP) derived from patients. The team then converts the extracted RNA to component DNA (cDNA), which is used for estimating viral loads by quantitative PCR (qPCR).
After performing qPCR, the researchers amplified the viral cDNA genome in 96-well format to reduce contamination from the sequences and to increase the replication of viral genomes. In this way, the researchers used two pools of primers designed by the ARTIC Network.
Afterwards, the team performed a complex PCR with two baths of ARTIC primers, and the two separate PCR output pools for each sample are combined, while the amplicons are separated by a cleaning step.
Finally, genomees with appropriate transmission can be seen with phylogenetic analyzes and variable call generated using web-based tools such as Nextstrain.
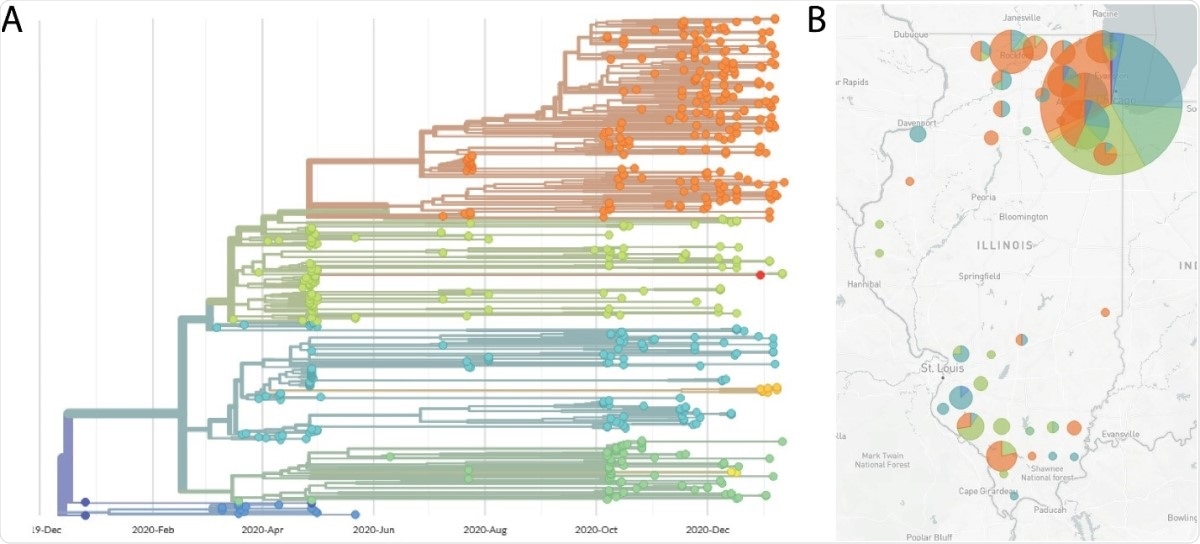
Nextstrain phylogenetic regression and map view of SARS-CoV-2 genome sequences. (A) A phylogenetic tree formed by the Nextstrain pipeline. Series come from samples taken in Illinois, between April 2020 and January 2021, and ordered by our lab. Each color represents a different Nextstrain cover. (B) Map of Illinois showing sample locations, by county. The size of the circle reflects the reasonable number of lines that came from that county. The pie chart shows the proportion of Nextstrain burrows in that location. The clade specification colors for panel A and B are: 19A (dark blue), 19B (blue), 20A (light blue), 20B (green), 20C (yellow-green), 20D (yellow), 20E (yellow-orange ), 20G (orange).
For this reason, the protocol can provide an easier way to sequence. The protocols implement ARTIC PCR and MinION nanopore to classify 96 samples simultaneously.
The team believes that the new protocol can provide a reliable starting point for those wishing to produce SARS-CoV-2 genome sequences using nanopore sequence technology.
* Important message
bioRxiv publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be seen as final, guiding health-related clinical practice / behavior, or be treated as information established.
Source:
Magazine Reference: