Bruker Corporation today announces a progressive publication from the professors Matthias Mann and Fabian Theis in the journal Nature Communications entitled ‘Deep learning the collisional crosss of the peptide universe from a million experimental values’ by Florian Meier et al. (doi.org/10.1038/s41467-021-21352-8).
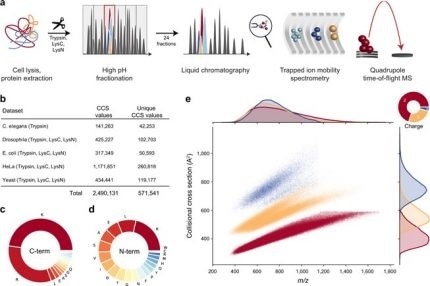
Figure 1. Large-scale peptide cross-section (CCS) measurement by TIMS and PASEF. From “In-depth study of the cross-sections of the peptide globe from a million experimental values”. (a) Workflow from the extraction of whole-cell proteomes through the digestion, breakdown and chromatographic separation of each fragment. The TIMS-rectangular TOF spectrometer was operated in PASEF mode. (b) Overview of the CCS dataset in this study by organization. (c) C-terminal peptide amino acid frequency. (d) N-terminal peptide amino acid frequency. (e) Distributed 559,979 unique data points, including modified series and cost state, in the CCS vs m / za color space according to the state code. Density distributions for m / z and CCS are projected on the upper and right axes, respectively. Source data is provided as a Source Data file. Image credit: Bruker Daltonics
The Nature Communication paper describes CCS values measured on the timsTOF Pro as the inherent property of the peptide ions, which can be used to increase confidence in peptide and protein group identification in gunpowder proteomics 4D. Since large proteomics based on spectrometry rely on matching spectra obtained correctly against a database of protein sequences, accurate CCS values offer the benefit of narrowing the list of candidates. This is essential for high sensitivity proteomics where low levels of peptide markers need to be accurately measured in complex mixtures, eg in plasma proteomics, peptidomics, immunopeptidomics or metaproteomics.
The publication summarizes a collaborative research effort led by Professor Matthias Mann, who holds dual positions at the Max Planck Institute of Biochemistry in Martinsried, Germany and the Novo Nordisk Foundation for Research Protein at the University of Copenhagen in Denmark, together with the group of Professor Fabian Theis, who also holds dual positions at the Helmholtz Munich Center in the German Research Center for Environmental Health, and in the TU Munich Department of Mathematics, in Germany.
The lead author, Dr. Florian Meier, now Associate Professor of Functional Proteomics at Jena University Hospital in Germany: “The scale and accuracy of CCS peptide values in our data from the timsTOF Pro were sufficient to properly train our in-depth learning model. index CCS values based solely on the peptide sequence. This connection between the amino acids contained within a peptide sequence and its measured CCS has the remarkable potential to increase protein recognition confidence. Since CCS peptide values are completely determined by their amino acid sequence sequences, they should be predicted with great accuracy and our in-depth learning model accurately predicted CCS values even for peptides. not previously served. We obtained data from full-protein scales of five organisms, which resulted in a volume of over two million CCS values, containing approximately 500,000 unique peptides, making it the most complete CCS dataset to date. seo. ”
The source code is publicly available to accelerate further developments for training and prediction models of the human peptide globe. Sensibly, our CCS model could make a god-PASEF® faster and cheaper by reducing the effort to generate libraries. In addition, CCS values intended to allow the use of community libraries, such as the Pan Human library, a source of more than 10,000 human proteins, should be used for targeted proteomics. ”
Matthias Mann, Professor
Dr Fabian Theis said: “In-depth learning, especially the recurring cloud networks used by many simulations, needs to be predictable, so I was delighted when Matthias into me and we were able to predict and differentiate the biochemical properties of peptides based solely on their order. I personally liked that we could therefore impose CCS values on many peptides that have never been measured. “
This paper demonstrates the tremendous potential of unambiguous CCS values for TIMS-PASEF methods in unbiased, in-depth 4D-ProteomicsTM. The proof strength, higher mobility and ultra-high sensitivity of the timsTOF platform are ideal for translational research. Large-scale peptide CCS values provide a fundamental advantage in protein recognition and measurement confidence in biomarker research in large cohort studies. In addition, the benefits of CCS values for enhancing cognitive confidence are also relevant to other timsTOF multiomics workflows, such as metabolomics, lipidomics and glycomics. These are exciting times for the fast growing timsTOF user community. ”
Dr. Gary Kruppa, Bruker Vice President, Proteomics
Source:
Magazine Reference:
Meier, F., Köhler, ND, Brunner, AD. et al. In-depth study of the cross-sections of the peptide globe from a million experimental values. Nat Commun 12, 1185 (2021). https://doi.org/10.1038/s41467-021-21352-8