The cells in our body are constantly fighting the risk of cancer by repairing damaged DNA. In a new study, scientists from Tokyo University of Science study the complex structure of accessible proteins that play a key role in the activation of the “Fanconi anemia pathway” involved in DNA repair, and report on the factors which governs its stability. Their ideas can help find novel cures for disorders including chromosomal instability, including cancer.
DNA repair is one of the most essential functions performed by the cells in our body, an action that is so vital to our well-being that it is able to perform it to lead to results as horrific as cancer. The process of DNA repair involves a complex interaction between several gene pathways and proteins.
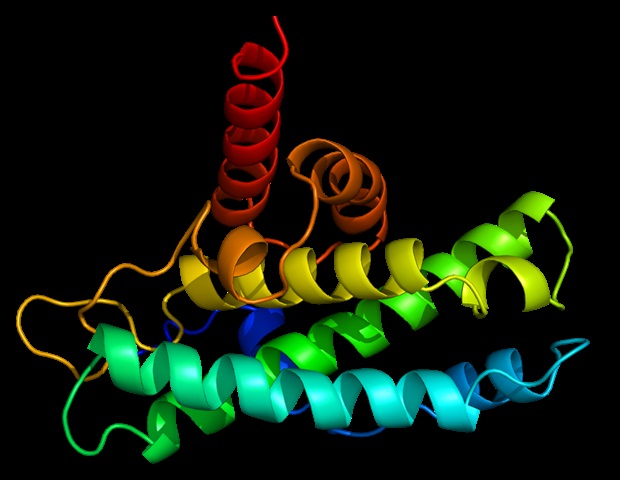
One such pathway is the “Fanconi anemia (FA) pathway,” in which genes are involved in DNA repair. FANCM, part of this pathway, is responsible for eliminating harmful “inter-strand cross-linking” of DNA and interacting with another component called MHF to function. The importance of FANCM-MHF complex is well documented: its loss can lead to chromosomal instability that can cause diseases such as FA itself and cancer. However, little is known about its structure and the basis of its sustainability.
Against this background, Associate Professor Tatsuya Nishino and his colleague Dr. Sho Ito of Tokyo University of Science decided to study the crystalline structure of this intricate complex using separation methods. X-ray.
DNA damage and chromosome segregation are essential tools for the maintenance and possession of genes that all organisms have. MHF (also known as CENP-SX) is an enigmatic center that plays a role in DNA repair and chromosome segregation. We wanted to find out how it performs these two different tasks in the hope that it will give us a glimpse of novel onions. “
Tatsuya Nishino, Associate Professor, Tokyo University of Science
Their findings are published in Acta Crystallographica Section F: Structural Biology Communication.
The scientists prepared a revised version of the FANCM-MHF facility, composed of FANCM from chickens and MHF1 and MHF2. They were able to purify three different types of protein crystals – tetrahedral, needle-shaped, and rod-shaped – from similar crystallization conditions. Surprisingly, when the structure was confirmed by X-ray crystals, they found that two of the crystal shapes (tetrahedral and needle-shaped) contained only the MHF complex without FANCM.
Annoyed by this discovery, the scientists used biochemical methods to study the causes of the FANCM-MHF complex. They were expressed as a result of a fertilizer called 2-methyl-2, 4-pentanediol (MPD), an organic solvent commonly used in crystals, and exposure to an oxidizing environment.
But, what exactly is the separation happening? The scientists believe that this may be caused in part by some unsafe amino acids in the chicken FANCM which causes the complex to accumulate with other FANCM-MHF complex and dismantle. In addition, they suggest that a small flexible MPD structure may have allowed FANCM to attach to and release, eliminating the build-up.
The findings are remarkable and can be used to improve the stability of the FANCM-MHF complex for future studies of its structure and function. Dr Ito believes that we have much to look forward to in the future from this complex. “A good understanding of this complex can help us treat cancer and genetic diseases, create artificial chromosomes, and even develop new biotechnological devices,” he thinks.
Thanks to the efforts of Professor Nishino and Dr Ito, we are already one step closer to that goal!
Source:
Tokyo University of Science
Magazine Reference:
Ito, S & Nishino, T (2021) Structural analysis of the FANCM – MHF chicken complex and its stability. Acta Crystallographica Section F: Structural Biology Communication. doi.org/10.1107/S2053230X20016003.