A team of HIV researchers, cell biologists, and biophysicists who came together to support the science of COVID-19 determined the atomic structure of a coronavirus protein that is thought to help avoid the pathogen and respond from human immune cells to reduce. The structural map – now published in the journal PNAS, but open to the scientific community since August – has laid the groundwork for new antiviral treatments designed specifically for SARS-CoV-2, and has allowed further studies on how the emergence of a new virus infects the human body.
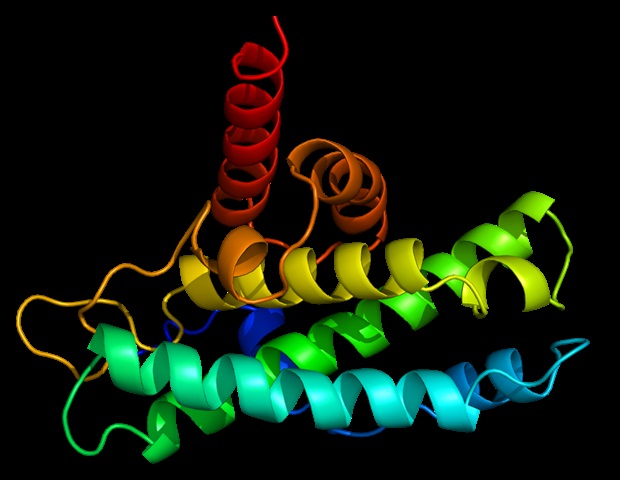
Using X-ray crystals, we constructed an atomic model of ORF8, and identified two specific regions: one that is only present in SARS-CoV-2 and its ancestor bats immediately, and one that is not present from SARS-CoV-2. any other coronavirus. These segments form the basis of the protein – which is a secretory protein, which is not bound to the membrane like the normal spike proteins of the virus – and creates a new intermolecular interface. We, and others in the research community, believe that these interface resources are involved in reactions that in some way make SARS-CoV-2 more pathogenic than the rays from which it originated. e. “
James Hurley, Lead Author, UC Berkeley Professor and Associate Faculty, Lawrence Berkeley National Laboratory (Berkeley Lab)
Structural biology under consideration
Labor is always mapping the structure of a protein, because scientists have to invent bacteria that can pump large amounts of the molecule, treat the molecules into a pure crystalline form, and then many, many separation images. Take an X-ray of the crystals. These images – taken as an X-ray beam kicking off atoms in the crystals and passing through gaps in the surface, generating a pattern of spots – are taken together and analyzed through special software to find out the whereabouts of each individual atom. Depending on the complexity of the protein, this tedious process can take years.
For many proteins, the process of map construction is aided by comparing the structure of the undissolved molecule with other proteins with similar amino acid sequences that have already been mapped, allowing -science make informed estimates of how the protein folds into its 3D shape.
But for ORF8, the team had to start from scratch. The amino acid sequence of ORF8 is so unlike any other protein that scientists have not reported its complete shape, and it is the 3D shape of a protein that proves its function.
Hurley and his colleagues UC Berkeley, with experience in structural analysis of HIV proteins, worked with Marc Allaire, a biologist and crystal specialist at the Berkeley Center for Structural Biology, located at Source Berkeley Lab Advanced Lighting (ALS). Together, the team worked in overdrive for six months – Hurley’s lab generated crystal samples and forwarded them to Allaire, which would use ALS’s X-ray beamlines to take the separation images. It yielded hundreds of crystals with multiple versions of the protein and thousands of diffraction images analyzed by special computer algorithms to bring together the structure of ORF8.
“Coronaviruses move differently than viruses such as the flu or HIV, which quickly accumulate many small changes through a process called hypermutation. In coronaviruses, large lumps of nucleic acid sometimes occur. moving around through repetition, “Hurley explained. When this happens, large, new sections of protein can appear. Genetic analyzes carried out very early in the SARS-CoV-2 pandemic revealed that this new strain had emerged from a bat-infecting coronavirus, and that mutation was re- an important factor occurring in the region of the genome encoding for a protein, called ORF7, found in many coronaviruses. The new form of ORF7, called ORF8, quickly gained the attention of virologists and epidemiologists because genetically differentiated events such as the one seen for ORF8 often cause new strain violence.
“Basically, this mutation caused the protein to double in size, and the doubling material was not associated with any known folding,” Hurley said. “A heart around half of it is associated with a well-known complex type in the release structure of earlier coronaviruses, but the other half was completely new.”
Answering the call
Like so many scientists working on COVID-19 research, Hurley and his colleagues chose to share their findings before the data could be published in a peer-reviewed journal, allowing with others beginning influential follow-up studies months earlier than the traditional publication process would have allowed. As Allaire explained, the therapeutic crisis caused by the pandemic shifted everyone in the research community to a pragmatic mindset. Instead of worrying about who achieved something in the first place, or sticking to their specific areas of study, scientists shared data early and often, and took on new projects when the resources and knowledge they needed.
In this case, the co-authors of Hurley UC Berkeley had the knowledge of viral and crystalline protein, and Allaire, a longtime collaborator, was up the slope, also with crystalline knowledge and, crucially, a railroad that still working. The ALS had received specific funding from the CARES Act to stay working for COVID-19 checks. The team knew from a review of SARS-CoV-2 genomic analysis posted in January that ORF8 was an important part of the (then much more dangerous) pandemic puzzle, so they started working .
Since then the authors have moved on to other projects, satisfied that they laid the foundation for other organizations to study ORF8 in more detail. (Currently, several studies are ongoing with a focus on how ORF8 interacts with cell receptors and how it interacts with antibodies, as infectious people appear to take out antibodies that bind to ORF8 as well as antibodies specific to the surface proteins of the virus.)
“When we started this, other projects were launched, and we had this unique opportunity to detect and solve an urgent problem,” said Allaire, who is part of the Department of Molecular Biophysics and Integrated Bioimaging. Berkeley Lab. “We worked closely, with a lot of back and forth, until we got it right. It has been one of the best collaborations of my career. “
Source:
Lawrence Berkeley National Laboratory
Magazine Reference:
Flour, TG, et al. (2021) Structure of SARS-CoV-2 ORF8, a rapidly evolving protein. PNAS. doi.org/10.1073/pnas.2021785118.